BSDB Gurdon Summer Studentship Report (5)
Posted by BSDB, on 2 December 2015
![]() In 2014, the British Society of Developmental Biology (BSDB) has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. Each year, 10 successful applicants spend 8 weeks in the research laboratories of their choices, and the feedback we receive is outstanding. Please, read the student report, kindly sent to us by Anna Klucnika who studies at Cambridge University and was hosted in summer 2015 by Aziz Aboobaker in Oxford.
In 2014, the British Society of Developmental Biology (BSDB) has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. Each year, 10 successful applicants spend 8 weeks in the research laboratories of their choices, and the feedback we receive is outstanding. Please, read the student report, kindly sent to us by Anna Klucnika who studies at Cambridge University and was hosted in summer 2015 by Aziz Aboobaker in Oxford.
Immortal worms
 The Gurdon Summer Studentship has allowed me to undertake a fascinating summer project in the laboratory of Aziz Aboobaker in Oxford, working with the planarian Schmidtea mediterranea, an exciting model for stem cells, ageing and cancer1. These flatworms have the capacity to regenerate completely from the smallest fragment from almost any body part, owing to the widespread abundance of neoblasts in their mesenchyme. Neoblasts, or at least a subset, have been demonstrated to be pluripotent by single cell injection into lethally irradiated worms resulting in complete rescue2.
The Gurdon Summer Studentship has allowed me to undertake a fascinating summer project in the laboratory of Aziz Aboobaker in Oxford, working with the planarian Schmidtea mediterranea, an exciting model for stem cells, ageing and cancer1. These flatworms have the capacity to regenerate completely from the smallest fragment from almost any body part, owing to the widespread abundance of neoblasts in their mesenchyme. Neoblasts, or at least a subset, have been demonstrated to be pluripotent by single cell injection into lethally irradiated worms resulting in complete rescue2.
Since vertebrates do not have adult stem cells that are pluripotent, planarians provide a unique opportunity to study the mechanisms of stem cell maintenance, induction of differentiation, whole body regeneration, as well cancer related stem cell behaviour.
An interesting feature of neoblasts is the presence of chromatoid bodies, which strikingly resemble germ granules found in germ cells across metazoa. These are electron dense structures made up of RNPs involved in posttranslational gene regulation in the germline. Chromatoid bodies and germ granules contain many homologous proteins, which suggests a conserved germline multipotency program3. Except for Nanos, neoblasts express most germ line specific genes, such as homologues to Bruli, Piwi, and Tudor, and RNAi depletion of these proteins blocks regeneration and indicates involvement in neoblast maintenance and differentiation4.
Does Tudor reign over pluripotency?
The focus of my project was on the Tudor homologue in S.mediterranea, Smedtud-1. RNAi in a related species, S. polychroa, results in neoblast depletion, indicating a role in long-term neoblast maintenance5. From studies in other organisms we know that the Tudor domains of Tudor bind symmetrically dimethylated arginines of Piwi proteins and glycolytic enzymes in the germ granules6,7. These studies and knockdowns indicate that Tudor is involved in piRNA synthesis required for gametogenesis and stem cell maintenance. The role that Tudor may have as a pluripotency factor made me keen to investigate this mysterious protein.
What makes the picture even more interesting is that RNP granules are also found in planarian neurons and Smedtud-1 is expressed in the CNS as well as in germ cells and neoblasts8,9. However it is not known in which specific neurons Tudor is expressed or when Tudor expression is switched on and/or off.
Elucidating the Tudor lineage
I wanted to find out what is the pattern of Tudor expression in stem cell progeny as they differentiate into neurons. To do this I carried out fluorescence in situ hybridization (FISH) for several markers (tudor, coe, th, tph, ChAT10) to visualize their expression on top of an immunostain for Tudor protein.
Coe is a marker for neural progenitors and neurons and so I synthesised antisense probes to detect coe to see whether Tudor is co-expressed continuously throughout the neuronal lineage or not.
To establish where in the CNS Tudor is expressed, various markers for specific neuronal subtypes were used- th for dopaminergic, tph for serotonergic and ChAT for cholinergic neurons. Anish, a DPhil student, was thus able to help me show that Tudor protein is not expressed, for example, in dopaminergic neurons but in proximal neurons.
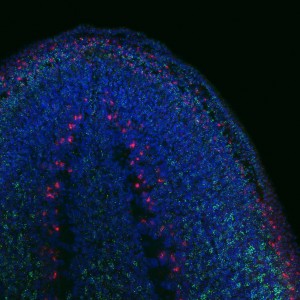
To find out whether the expression pattern of the transcript matches that of Tudor protein. I used the genome data available to clone Smedtud-1 to synthesise antisense probes that I used for FISH jointly with immunostaining for the protein using the Tudor antibody.
Although I wasn’t able to collect all of my data in the short time that I was in the lab, I’ve achieved so much. I’ve learnt how to cut worms, microinject, clone genes, synthesise probes and carry out immuno and in situ protocols. I’ve learnt to always ask when in doubt. I’ve learnt to be scrupulous. I’ve learnt that it can be very frustrating when experiments don’t work out as planned and when time runs out. But those little setbacks showed me just how determined I am to do science.
The Aboobaker lab was extremely welcoming and supportive. Thank you to Aziz, Natasha, Dani, Nobu, Prasad, Anish, Sounak, Yuli, Sam, Damian, Alvina, Holly, Ben and Alex.


 (7 votes)
(7 votes)