BSDB Gurdon Summer Studentship Report (7)
Posted by BSDB, on 4 December 2015
![]() In 2014, the British Society of Developmental Biology (BSDB) has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. Each year, 10 successful applicants spend 8 weeks in the research laboratories of their choices, and the feedback we receive is outstanding. Please, read the student report, kindly sent to us by Isabella Watts(Oxford University) who was hosted in summer 2015 by Tatjana Sauka-Spengler at the Weatherall Institute of Molecular Medicine in Oxford..
In 2014, the British Society of Developmental Biology (BSDB) has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. Each year, 10 successful applicants spend 8 weeks in the research laboratories of their choices, and the feedback we receive is outstanding. Please, read the student report, kindly sent to us by Isabella Watts(Oxford University) who was hosted in summer 2015 by Tatjana Sauka-Spengler at the Weatherall Institute of Molecular Medicine in Oxford..
A transcriptional analysis of the innate immune response to melanoma: the promise of the zebrafish
 This summer I was privileged enough to receive a Gurdon Summer Studentship to work for 8 weeks in the laboratory of Tatjana Sauka-Spengler, at the Weatherall Institute of Molecular medicine. About to go into my third year of studying medicine at Oxford, I have spent the majority of the first two years of my degree learning about the science that underpins medicine. All our lecturers highlight the importance of understanding the scientific evidence that underlies the concepts that we learn. However, no matter how much time you spend trawling through the depths of PubMed to find a new and exciting paper for an essay, it’s not the same as getting hands on experience in a laboratory and learning about different scientific techniques.
This summer I was privileged enough to receive a Gurdon Summer Studentship to work for 8 weeks in the laboratory of Tatjana Sauka-Spengler, at the Weatherall Institute of Molecular medicine. About to go into my third year of studying medicine at Oxford, I have spent the majority of the first two years of my degree learning about the science that underpins medicine. All our lecturers highlight the importance of understanding the scientific evidence that underlies the concepts that we learn. However, no matter how much time you spend trawling through the depths of PubMed to find a new and exciting paper for an essay, it’s not the same as getting hands on experience in a laboratory and learning about different scientific techniques.
My project involved helping my supervisor Amy Kenyon with the final steps of her PhD. Amy had generated a transgenic zebrafish model to study the innate immune response under conditions of inflammation, including tuberculosis and melanoma. This model system harnesses developmental biology and basic science to provide an invaluable tool to better understand the mechanisms of different disease states.
I was mainly focussing on the innate immune response to melanoma, which is an incredibly exciting area of research as macrophages and neutrophils form a major part of the tumour microenvironment. These cells have diverse roles in tumours and can both destroy transformed cells and also contribute to tumour development. However, it has proven difficult to fully examine the interactions of these cells and transformed cells in vivo due to limitations in current model systems. Amy developed a unique model system, pioneered in vivo by the Sauka-Spengler laboratory, which allows isolation of specific cell types and cellular compartments directly from the organism, via a genetically encoded, tissue-specific biotin tagging system.
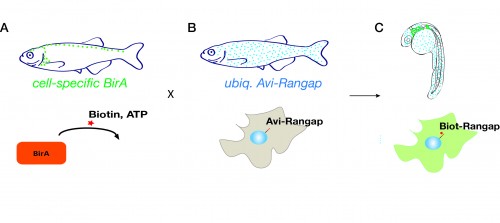
The system involves fish that ubiquitously express the nuclear envelope-associated protein Rangap fused to an Avi-tag (Avi-Rangap). These have then been crossed with a separate transgenic line where E.coli biotin ligase (BirA) is selectively expressed under control of the mpeg or mpo promoter which are specifically active in macrophages or neutrophils, respectively. Therefore, in the macrophages or neutrophils, biotin is covalently added to the Avi-tag Rangap protein, resulting in the ‘biotinylation’ and tagging of the nuclei of these cells, which can then be isolated using highly stringent streptavidin-biotin affinity purification protocol.
To generate an inducible melanoma model, another binary system of transgenic fish has been used. In the driver line, the bacterial LexPR repressor is fused to a truncated form of the human progesterone receptor, all under the control of a melanocyte specific promoter. The effector line harbours the Lex Operon, controlling the expression of a Ras-oncogene fused to an mCherry fluorescent reporter. When the synthetic steroid mifepristone is added to the water of embryos carrying both alleles, it binds to the LexPR transactivator which, in turn, binds to the Lex Operon region and drives expression of Ras-oncogene specifically in melanocytes, resulting in their transformation and proliferation.
From the very first day of my project I learned a huge amount, and the learning curve was definitely vertical at times! I was incredibly lucky to be working on a very broad topic which meant that I was able to try my hand at a huge number of different techniques.
Some of the earliest work I did involved confirmation of the different model lines I was using. In my first experiment, I wanted to check whether the melanoma model we were using effectively mimicked the cancer over-proliferation phenotype. In the zebrafish embryo melanocytes are usually post-mitotic at 60 hours post fertilisation, thus I wanted to confirm that induction of our cancer system resulted in continued cell proliferation past this point. We injected EdU, a thymidine analogue incorporated into the DNA of dividing cells, into the pericardium of day 4 embryos, in order to label proliferating cells. Upon imaging we saw that, in fish where the LexPR system was activated, there was continued proliferation of melanocytes whilst in control fish there was no overlap between the melanocytes and the stain that labelled the dividing cells.
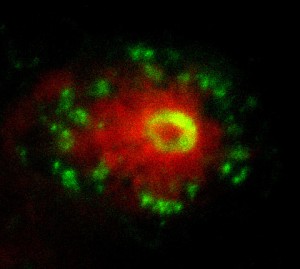
In the next step of my project I made use of the transparency of the early zebrafish embryo to image the live interaction between the neutrophils, macrophages and transformed melanocytes (Fig. 2). The final step of my project was a transcriptional analysis of the interactions between the innate immune cells and tumour cells. Collecting day 5 embryos where we had induced the oncogenic transformation in melanocytes, we selectively isolated the nuclei of macrophages and neutrophils from the fish using the streptavidin-biotin affinity purification previously mentioned. Whilst incredibly rewarding, this did involve spending entire days in the cold room, and I got a lot of strange looks as I carried 3 jumpers into work in the blazing sun and even drank hot chocolate in the institute’s cafeteria on the hottest day of the year! After isolating the RNA from the nuclei and making cDNA libraries, the data was sent off for sequencing and we were able to see which genes were up- and down-regulated in these cells in response to the cancer.
Looking back at my time in the lab I can’t quite believe how much I have learned! I have had an amazing time and every week has been packed with new things. I loved getting to learn more about the processes involved in laboratory science and I have been convinced that in the future I want to combine both clinical work and research. This has been a very valuable experience and I would really encourage other university students to apply for a Gurdon Summer Studentship. I also want to say a huge thank you to everyone in my lab, especially Tatjana and Amy for making my summer so interesting!
Isabella Watts


 (5 votes)
(5 votes)