BSDB Gurdon/The Company of Biologists Summer Studentship Report #26 – Courtney Lancaster
Posted by BSDB, on 18 December 2018
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding. You can read accounts from previous years here.
Our fifth report from the 2018 group of student awardees comes from Courtney Lancaster (student at Durham) who undertook her research with Ben Steventon (University of Cambridge).
Deciphering the dynamics of neuromesodermal progenitors at the end of axis elongation in the zebrafish embryo: A tail of a summer research project
I was introduced to developmental biology in the second year of my undergraduate degree at Durham University. From the first lecture, I was amazed and I left the lectures with more questions than answers on many occasions. This curiosity stemmed my quest to find a lab where I could begin to answer some of my questions. Of particular interest to me is the elongation of the vertebrate body axis, with a key question: How does the body axis stop elongating?
I was lucky enough to come across the Steventon lab, who focus their research on a population of bipotent stem cells called neuromesodermal progenitors (NMps). NMps co-express Sox2 and Brachyury (neural and mesoderm markers respectively) and they provide progenitor cells for the spinal cord and mesoderm during vertebrate axis elongation (Steventon and Martinez-Arias., 2017). Furthermore, NMps are a conserved source of spinal cord amongst vertebrate species making comparative studies to be of great interest (Steventon et al., 2016).
In mouse and chick embryos, NMps are a population of bipotent, self-renewing progenitor cells whose derivatives undergo a great deal of cell proliferation that is one of the main drivers of axis elongation. On the other hand, zebrafish body axis extension is more a product of cell movements rather than of volumetric growth (Steventon et al., 2016). Therefore, a key question is: To what degree do NMps self-renew in the zebrafish embryo to give rise to both neural and mesodermal cell fates? To begin answering this question, I carried out lineage tracing at single-cell resolution from a light-sheet movie of the zebrafish tail. This analysis meant that I got to grips with embryo mounting and I worked closely with computer scientists and engineers from the Cambridge Advanced Imaging Centre (CAIC). I was lucky to have access to a light-sheet microscope in which the stage position continually corrected to follow the tail. This allowed the tail to stay in view whilst the embryo was undergoing development, which is essential for long-term imaging of an elongating structure.
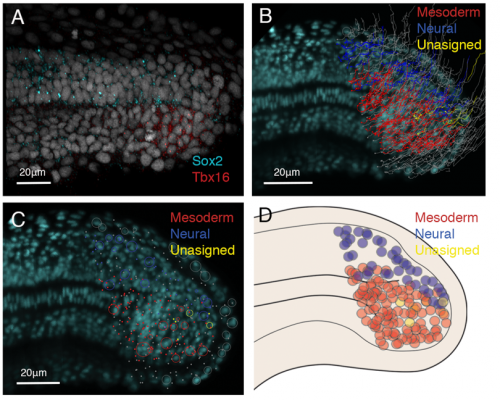
From the light-sheet dataset, all nuclei were segmented and automatically tracked using TGMM tracking software (Amat et al., 2014). To select my area of interest, the tailbud cells, computer scientists at CAIC wrote a Matlab script allowing me to select the area I wanted to track. I was then able to manually correct the tracks of interest using a Fiji plug in called MaMuT (Figure 1B) and assign fates according to gene expression data (figure 1A). The cells were tracked from 21 somite stage to the completion of somitogenesis, the stages where NMps contribute to the final stages of the body axis (Steventon and Martinez-Arias., 2017). From this data we concluded that little cell division occurs in the NMp population and that NMps are mono-fated progenitor cells, either giving rise to the neural or mesodermal lineages, but not both (Figure 1B, C, D). Therefore, in zebrafish, NMps do not undergo vast amounts of cell division to continually contribute to the elongating body axis.
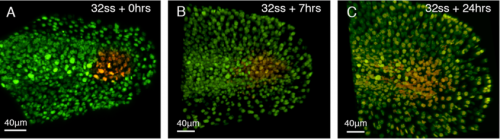
Apoptosis takes place at the end of axis elongation in chick embryos. This occurs following a rise in retinoic acid signaling and the loss of FGF dependent mesoderm identity (Olivera-Martinez., 2012). Considering the different cell behaviors and contribution of NMps to the body axis in zebrafish, I next asked the question: Does apoptosis have a role in terminating the body axis in zebrafish embryos? To gain insight into this, cells at the end of the presomitic mesoderm (PSM) were photo-labelled by injecting embryos, at one cell stage, with a photoconvertible fluorescent protein mRNA called Kikume. A confocal microscope was then used to shine UV light onto the posterior PSM to convert the cells from green to red (figure 2A) and these cells could then be followed over time (figure 2B, C). It was found that the labelled cells did not noticeably undergo apoptosis and they did not segment (figure 2). It will be essential to confirm this finding with antibody immuno-labelling of apoptosis associated proteins such as caspase 3 or with a TUNEL assay. Nonetheless, morphological analysis suggests that apoptosis does not precede the termination of axis elongation in zebrafish embryos.
As well as the two main projects above, I also began to analyse the light-sheet data to understand which cell movements are responsible for elongating the body axis. This was carried out on Imaris software, which allowed me to view the tailbud in 3D and to select different tissues (e.g. PSM) for analysis. Further to this, I used photolabelling (method previously described) to look at the contribution of different tailbud progenitor populations to the PSM and somites.
This experience has been invaluable to me and I have thoroughly enjoyed mixing lab experiments with computational analysis, which are both important in science.
My interaction with academics from CAIC made me realize the importance of interdisciplinary science in order to make scientific research more productive. It allows biologists to gain better insights as well as improving computational techniques for the field as a whole. I have also attended lab meetings and had the opportunity to present my work. Further to this, I attended a fantastic conference “Engineering Multicellular Self-Organisation III”. When I thought that my summer could not get any better, my light-sheet data and analysis presented here has been accepted as part of a research article in the Development journal. Together, the skills gained here have taught me to be more critical and have begun to equip me for an exciting career in research.
I would like to thank Ben Steventon for making this incredible experience possible and for the great advice and discussions throughout the project. Thank you to the Steventon group: Lewis, Tim, Kane, Susie, Berta, Carolina, and John for creating a welcoming and enthusiastic working environment, not forgetting the tea breaks and pub nights! Last but not least, thank you to the BSDB Gurdon Studentship for financial support.
References
Amat F, Lemon W, Mossing, D. P, McDole K., Wan Y, Branson K, Myers E. W, Keller, P. J. (2014). Fast, accurate reconstruction of cell lineages from large scale fluorescence microscopy data. Nat. Methods 11: 951–958.
Olivera-Martinez I, Harada H, Halley PA, Storey KG (2012) Loss of FGF-Dependent Mesoderm Identity and Rise of Endogenous Retinoid Signalling Determine Cessation of Body Axis Elongation. PLoS Biol 10(10): e1001415.
Steventon, B., Duarte, F., Lagadec, R., Mazan, S., Nicolas, J.-F. and Hirsinger, E. (2016). Species tailoured contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143(10): 1732–41.
Steventon, B. and Martinez Arias, A. (2017). Evo-engineering and the cellular and molecular origins of the vertebrate spinal cord. Dev. Biol. 432(1): 3-13.




 (No Ratings Yet)
(No Ratings Yet)