From basic questions to exciting findings
Posted by Hadi Boukhatmi, on 21 May 2018
In this post we report the backstories behind our recently published paper. It was an enjoyable research adventure driven by discussions, readings, exciting experiments and unexpected discoveries. As a result, we described a novel molecular mechanism underpinning stem cell and progenitor maintenance during development. Here is the sequence of the main events that inspired us in making this story.
The foundations; it’s good to have a side project
The project has started from the following basic developmental biology question: How are the Drosophila flight muscle progenitors (MPs) kept undifferentiated throughout much of the animal development? To solve this question, we initially embarked on a genome wide approach to define their unique characters. But after many rounds of dissections and ChIPs, the ChIP-seq results that came back (over the Christmas break!) they were basically meaningless and we were lucky that we had a side project in parallel with one of our favourite genes. Our enthusiasm for the zinc finger homeodomain transcription factor Zfh1/ZEB came from its identification in a previous genome wide screen for direct Notch targets in different Drosophila cell lines, including Drosophila DmD8 cells (MP-like cells) (Krejcí et al., 2009). Also, it was well established that Zfh1, as well as its mammalian homologues ZEB1/2, had roles in maintaining stem cells. Together these elements prompted us to investigate the role of zfh1 in MPs maintenance and we ended up showing that indeed Notch pathway activates directly zfh1 expression in vivo in the MPs and this cascade is required to keep them undifferentiated. This regulatory mechanism set the basic foundations for our subsequent work.
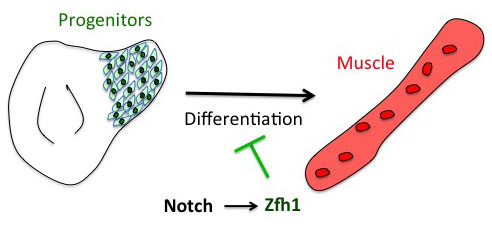
How the microRNA miR-8 entered the scene
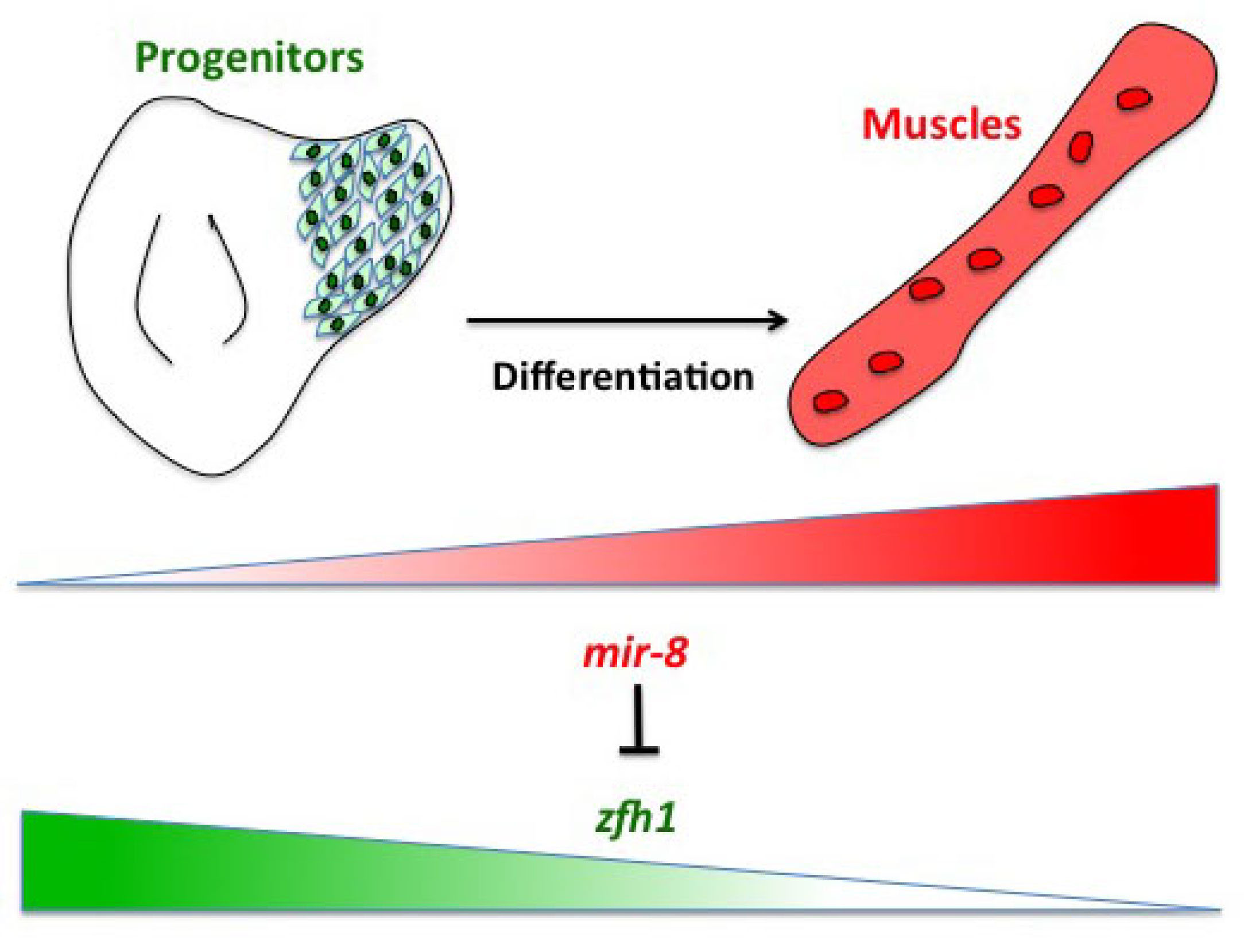
Having described the Notch/Zfh1 regulation, we realised that one important question was how the progenitors cells get rid of Zfh1 in order to differentiate. This led us to a paper from Maria Dominguez lab, which linked zfh1 to the micro RNA miR-8 in the intestinal stem cells (Antonello ZA et al., 2015). Indeed we discovered from there that miR-8/zfh1 regulation orchestrates many developmental process in either Drosophila or vertebrates so it seemed natural to check whether it plays any role in the muscle differentiation program … as turns out to be the case. We found that miR-8 down regulated zfh1 in the MPs to facilitate the transition into functional flight muscles. Thus, we added miR-8 to the Notch/Zfh1 regulation and started to fill the gaps, strengthen the data and think about how to complete the story.
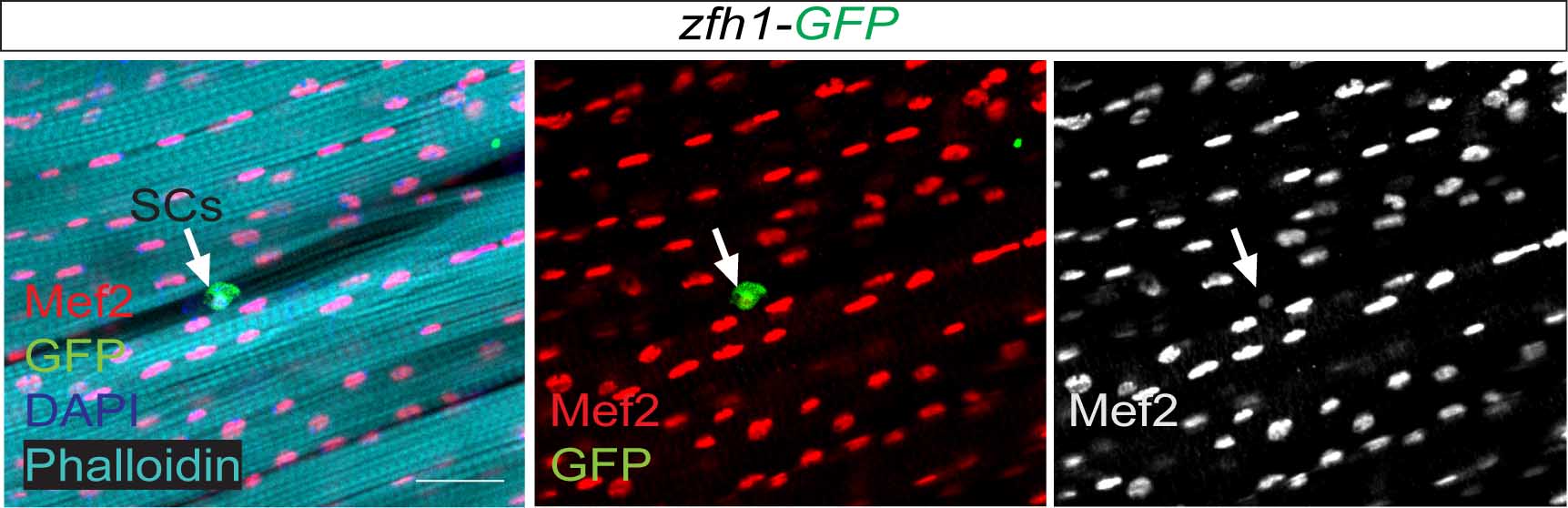
Satellite cells do exist in Drosophila ?
So far the focus was on the muscle progenitors prior to the muscle differentiation. But might some of these cells persist in the adult? At that time, I had no idea of how to do antibody stainings on adult muscles. Thankfully, Hannah Green and Juanjo Perez-Moreno kindly agreed to show me how to dissect and stain adults and pupal muscles. Excitingly there were Zfh1-positive cells associated with the adult muscles. But this did not immediately mean they were persistent muscle precurors…. the lab (aka my PI!) was not convinced! They pointed out that blood cells were also likely to be cruising around and that the zfh1 enhancer I used overlapped with a powerful blood cell enhancer identified by Alex Stark group (Zabidi M A et al., 2015). So next, I was tasked with double staining with blood cell markers to rule out this possibility. After a few trials I could finally show that indeed there was a population of cells expressing Zfh1 that were distinct from the plasmatocyte blood cells. So it seemed I was right! There is a persistent muscle progenitor cell population in adult flies similar to the satellite cells in mammals.
Very excitingly, another group had reached the same conclusions. In early 2016, we came across an intriguing pre-print paper from VijayRaghavan’s Lab describing muscle Satellite Cells (SCs) in adult flies (Now published as Chaturvedi D et al., 2017). In this manuscript the authors described and characterised a SC population, resident beneath the basal lamina of the adult muscles, and had found that they are lineal descendant of the larval MPs. So we carried out some similar lineage analysis to confirm that indeed the Zfh1-expressing cells had the same properties as their satellite cells. Furthermore, we could show, using one specific zfh1 enhancer to drive expression of a pro-apoptotic gene, that their elimination affected muscle homoestasis. Good news never comes alone: while we were writing our paper, an updated version of the VijayRaghavan Lab SC pre-print was released, they included new data with Zfh1 stainings in the SCs. It was reassuring to see that our data converged together!
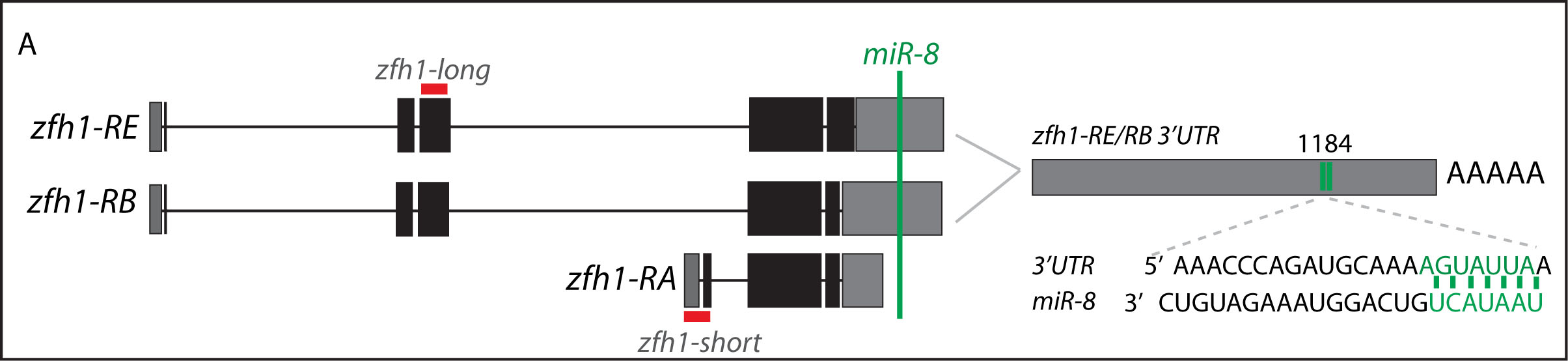
Short vs Long zfh1 isoforms: Why does length matter?
The discovery of these Zfh1-SCs in the adult flies placed us in an unprecedented position to address the exiting question of how such SCs escape the differentiation program during muscle development. Indeed, It is quite intriguing that this small subset of MPs behaves differently from the rest of the main pool of myoblasts that normally initiate the muscle differentiation program. In a discussion after my lab meeting presentation we ended up looking at the different zfh1 transcripts on Flybase (The online Drosophila database) and realised that there is a second zfh1 isoform, which is characterised by a shorter 3’UTR. Importantly, and unlike zfh1-long isoforms, zfh1-short isoform lacks the miR-8 seed site so it could be invisible to miR-8 detection. We thought this could be an exciting mechanism to protect the SCs and so we set out to test the hypothesis that a switch in zfh1 isoforms could be the trick to prevent SCs from differentiation. We quickly designed RNA-probes and checked the expression profile (by smFISH) of these isoforms during development. The results were striking and consistent with the initial hypothesis, since we found that zfh1-short is accumulated to high level specifically in the SCs. A series of experiments followed up and the manuscript started to have a pretty nice shape especially after the first round of elife reviewing process where the reviewers gave us many valuable comments and suggestions.
Conclusion
Our voyage ended in a very different place than we started it! Key to its success was challenging ourselves to go beyond the simple results and seizing the ideas from the literature. Bringing these together meant that we could delve into the mechanisms, but only with the additional inspiration from the data in Flybase did we make the final leap, which shows how important these resources are! In the end, we discovered a mechanism that explains how stem cells can escape the differentiation program and survive until the adult stage where they contribute to tissue homeostasis. At the root of it is a switch in RNA isoforms to produce RNAs that are insensitive to miRs. We believe that this is a powerful mechanism that may be of widespread significance.
References
Krejcí A, Bernard F, Housden BE, Collins S, Bray SJ. 2009. Direct response to Notch activation: signaling crosstalk and incoherent logic. Science Signaling.
Antonello ZA, Reiff T, Ballesta-Illan E, Dominguez M. 2015. Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8-Escargot switch. The EMBO Journal.
Chaturvedi D, Reichert H, Gunage RD, VijayRaghavan K. 2017. Identification and functional characterization of muscle satellite cells in Drosophila. eLife.
Zabidi MA, Arnold C D, Schernhuber K, Pagani M, Rath M, Frank O and Stark A. 2015. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature.


 (4 votes)
(4 votes)