In Development this week (Vol. 141, Issue 20)
Posted by Seema Grewal, on 7 October 2014
Here are the highlights from the current issue of Development:
Small (molecule) steps to making bone
 The repair of cartilage and bone following damage remains a clinical challenge. Current cell-based therapies rely mostly on adult mesenchymal stromal cells, but the expansion of these into correctly differentiated and functionally competent chondrocytes, which give rise to cartilage and then bone, remains problematic. Here, Naoki Nakayama and colleagues develop a small molecule-based approach that mimics the embryonic somitic chondrogenesis programme and can be used to differentiate mouse embryonic stem cells (ESCs) into chondrocytes in vitro (p. 3848). The authors first show that activation of Wnt signalling using a small molecule inhibitor of Gsk3 (CHR99021), together with inhibition of BMP signalling using a BMP type I receptor inhibitor (LDN193189), is sufficient to induce ESCs to form paraxial mesoderm-like progeny. This population, they report, expresses trunk paraxial mesoderm and somite markers but fails to express markers of sclerotome, which gives rise to cartilage. However, knowing that sonic hedgehog (Shh) and the BMP antagonist noggin are required for sclerotome induction in vivo, the researchers then demonstrate that short-term treatment of the mesodermal progeny with an Shh receptor agonist (SAG1) and the BMP inhibitor LDN193189 results in a sclerotome-like intermediate, leading to functional chondrocyte formation. When ectopically transplanted into immunocompromised mice, these chondrocytes were able to mineralise and form pieces of bone that contain marrow. This readily scalable and chemically defined method for directing chondrogenesis thus offers a promising approach for cartilage-mediated bone regeneration.
The repair of cartilage and bone following damage remains a clinical challenge. Current cell-based therapies rely mostly on adult mesenchymal stromal cells, but the expansion of these into correctly differentiated and functionally competent chondrocytes, which give rise to cartilage and then bone, remains problematic. Here, Naoki Nakayama and colleagues develop a small molecule-based approach that mimics the embryonic somitic chondrogenesis programme and can be used to differentiate mouse embryonic stem cells (ESCs) into chondrocytes in vitro (p. 3848). The authors first show that activation of Wnt signalling using a small molecule inhibitor of Gsk3 (CHR99021), together with inhibition of BMP signalling using a BMP type I receptor inhibitor (LDN193189), is sufficient to induce ESCs to form paraxial mesoderm-like progeny. This population, they report, expresses trunk paraxial mesoderm and somite markers but fails to express markers of sclerotome, which gives rise to cartilage. However, knowing that sonic hedgehog (Shh) and the BMP antagonist noggin are required for sclerotome induction in vivo, the researchers then demonstrate that short-term treatment of the mesodermal progeny with an Shh receptor agonist (SAG1) and the BMP inhibitor LDN193189 results in a sclerotome-like intermediate, leading to functional chondrocyte formation. When ectopically transplanted into immunocompromised mice, these chondrocytes were able to mineralise and form pieces of bone that contain marrow. This readily scalable and chemically defined method for directing chondrogenesis thus offers a promising approach for cartilage-mediated bone regeneration.OTX2 gets a head start
 The gene orthodenticle homologue 2 (Otx2) encodes a paired-type homeodomain transcription factor that is known to play a role in head morphogenesis. In the mouse, Otx2 is expressed in the anterior neurectoderm, where it is required for the differentiation of anterior neural tissues. Otx2 is also expressed in the anterior mesendoderm (AME) but its role here is unknown. On p. 3859, Patrick Tam and co-workers investigate the role of Otx2 in the AME. Using Otx2 AME conditional knockout embryos, the researchers show that Otx2 activity in the AME is essential for head formation. They further demonstrate that the expression of Dkk1 andLhx1, which are known regulators of head formation, is impaired in the AME of the Otx2conditional knockout embryos. Dkk1 is a direct target of Otx2, and the researchers further identify regulatory regions in the Lhx1 locus to which Otx2 can bind, suggesting that Lhx1 is also likely to be a direct target of Otx2. Finally, the analysis of AME-specific Otx2;Lhx1 andOtx2;Dkk1 compound mutant embryos reveals that Otx2 acts synergistically with Lhx1 andDkk1 in the AME during head formation. In summary, these findings uncover a crucial role for Otx2 during head and forebrain development.
The gene orthodenticle homologue 2 (Otx2) encodes a paired-type homeodomain transcription factor that is known to play a role in head morphogenesis. In the mouse, Otx2 is expressed in the anterior neurectoderm, where it is required for the differentiation of anterior neural tissues. Otx2 is also expressed in the anterior mesendoderm (AME) but its role here is unknown. On p. 3859, Patrick Tam and co-workers investigate the role of Otx2 in the AME. Using Otx2 AME conditional knockout embryos, the researchers show that Otx2 activity in the AME is essential for head formation. They further demonstrate that the expression of Dkk1 andLhx1, which are known regulators of head formation, is impaired in the AME of the Otx2conditional knockout embryos. Dkk1 is a direct target of Otx2, and the researchers further identify regulatory regions in the Lhx1 locus to which Otx2 can bind, suggesting that Lhx1 is also likely to be a direct target of Otx2. Finally, the analysis of AME-specific Otx2;Lhx1 andOtx2;Dkk1 compound mutant embryos reveals that Otx2 acts synergistically with Lhx1 andDkk1 in the AME during head formation. In summary, these findings uncover a crucial role for Otx2 during head and forebrain development.A new TALE of PU.1 function
 Numerous transcription factors (TFs), including PU.1 and Scl, are known to play important roles during haematopoiesis, but how these act within wider TF networks is unclear. Now, Berthold Göttgens and colleagues use transcription activator-like effectors (TALEs) to manipulate the expression of PU.1 and Scl and determine how these TFs function during developmental haematopoiesis (p. 4018). They first show that the modulation of PU.1 expression affects cell fate decisions during embryoid body haematopoiesis; PU.1 upregulation, for example, drives haematopoietic commitment but causes a loss of proliferative ability, whereas PU.1 repression inhibits the maturation and differentiation of early haematopoietic cells. They further report, using single-cell gene expression analyses, that TALE-induced PU.1 expression is associated with changes in the expression of several other haematopoietic genes, suggesting that early activation of PU.1 expression drives a haematopoietic programme at the expense of endothelial gene expression. Following on from this, the researchers show that the PU.1-14kb enhancer is active in the mid-gestation dorsal aorta in vivo, and that PU.1 is detectable in the early haemogenic endothelium. Together, these studies uncover a novel role for PU.1 during haematopoietic specification and highlight the use of TALEs in understanding developmental TF networks.
Numerous transcription factors (TFs), including PU.1 and Scl, are known to play important roles during haematopoiesis, but how these act within wider TF networks is unclear. Now, Berthold Göttgens and colleagues use transcription activator-like effectors (TALEs) to manipulate the expression of PU.1 and Scl and determine how these TFs function during developmental haematopoiesis (p. 4018). They first show that the modulation of PU.1 expression affects cell fate decisions during embryoid body haematopoiesis; PU.1 upregulation, for example, drives haematopoietic commitment but causes a loss of proliferative ability, whereas PU.1 repression inhibits the maturation and differentiation of early haematopoietic cells. They further report, using single-cell gene expression analyses, that TALE-induced PU.1 expression is associated with changes in the expression of several other haematopoietic genes, suggesting that early activation of PU.1 expression drives a haematopoietic programme at the expense of endothelial gene expression. Following on from this, the researchers show that the PU.1-14kb enhancer is active in the mid-gestation dorsal aorta in vivo, and that PU.1 is detectable in the early haemogenic endothelium. Together, these studies uncover a novel role for PU.1 during haematopoietic specification and highlight the use of TALEs in understanding developmental TF networks.Modelling morphogen-controlled gene expression
 Pattern formation during development often depends on the differential regulation of gene expression in response to a morphogen gradient, but how such gradients govern gene expression is unclear. A simplified view suggests that the morphogen activates a transcriptional activator, and that differential gene expression is dependent on the affinity or number of binding sites for this activator within target genes. However, this model does not account for bifunctional transcriptional effectors – those that function as activators and repressors – and has also been questioned by recent experimental results. Here, James Briscoe and colleagues describe a unifying mathematical model of morphogen-dependent gene expression that can explain recent counterintuitive findings (p. 3868). Using sonic hedgehog (Shh)-dependent patterning of the mouse neural tube as an example, the researchers develop mathematical models, based on statistical thermodynamic principles, that account for competitive binding of the active and repressive isoforms of Gli, the transcriptional effector of Shh, and that also represent other inputs that are known to regulate Shh target gene expression. Their modelling predicts that, for each Gli target gene, there is a neutral point in the Shh gradient, either side of which altering Gli binding affinity has the opposite effect on gene expression. They further report that inputs other than the morphogen determine the transcriptional response. Together, these analyses help reconcile conflicting results in the field and provide a theoretical framework that can be used to examine differential gene expression in other contexts.
Pattern formation during development often depends on the differential regulation of gene expression in response to a morphogen gradient, but how such gradients govern gene expression is unclear. A simplified view suggests that the morphogen activates a transcriptional activator, and that differential gene expression is dependent on the affinity or number of binding sites for this activator within target genes. However, this model does not account for bifunctional transcriptional effectors – those that function as activators and repressors – and has also been questioned by recent experimental results. Here, James Briscoe and colleagues describe a unifying mathematical model of morphogen-dependent gene expression that can explain recent counterintuitive findings (p. 3868). Using sonic hedgehog (Shh)-dependent patterning of the mouse neural tube as an example, the researchers develop mathematical models, based on statistical thermodynamic principles, that account for competitive binding of the active and repressive isoforms of Gli, the transcriptional effector of Shh, and that also represent other inputs that are known to regulate Shh target gene expression. Their modelling predicts that, for each Gli target gene, there is a neutral point in the Shh gradient, either side of which altering Gli binding affinity has the opposite effect on gene expression. They further report that inputs other than the morphogen determine the transcriptional response. Together, these analyses help reconcile conflicting results in the field and provide a theoretical framework that can be used to examine differential gene expression in other contexts.PLUS…
The T-box gene family: emerging roles in development, stem cells and cancer
 The T-box family of transcription factors exhibits widespread involvement throughout development in all metazoans. Here, Virginia Papaioannou provides an overview of the key features of T-box transcription factors and highlights their roles and mechanisms of action during various stages of development and in stem/progenitor cell populations. See the Primer on p. 3819
The T-box family of transcription factors exhibits widespread involvement throughout development in all metazoans. Here, Virginia Papaioannou provides an overview of the key features of T-box transcription factors and highlights their roles and mechanisms of action during various stages of development and in stem/progenitor cell populations. See the Primer on p. 3819
New insights into the maternal to zygotic transition
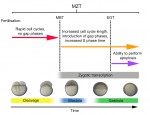 The initial phases of embryonic development occur in the absence of de novo transcription and are instead controlled by maternally inherited mRNAs and proteins. Following this period of transcriptional silence, zygotic transcription begins, the maternal influence on development starts to decrease, and dramatic changes to the cell cycle take place. Here, Steven Harvey and colleagues discuss recent work that is shedding light on the maternal to zygotic transition. See the Review on p. 3834
The initial phases of embryonic development occur in the absence of de novo transcription and are instead controlled by maternally inherited mRNAs and proteins. Following this period of transcriptional silence, zygotic transcription begins, the maternal influence on development starts to decrease, and dramatic changes to the cell cycle take place. Here, Steven Harvey and colleagues discuss recent work that is shedding light on the maternal to zygotic transition. See the Review on p. 3834


 (No Ratings Yet)
(No Ratings Yet)