Lifemap: a zoomable interface for exploring the entire Tree of Life
Posted by damiendevienne, on 30 January 2017
“Nothing in biology makes sense except in the light of evolution” (Dobzansky, 1973).
Our knowledge of the evolutionary relationships between all known organisms, the so-called Tree of Life (ToL), is crucial in all fields of biology. Many researchers in evolutionary biology are working on improving the quality and the comprehensiveness of this ToL. This implies better species sampling, more sequencing, improved evolutionary models and new algorithms for phylogenetic tree reconstruction. But displaying this information in a clear and simple way that is also convenient and useful to biologists outside the field of evolutionary biology has been largely underexplored.
In a paper published last December in PLOS Biology (de Vienne, 2016), I described an online tool, called Lifemap, for exploring the entire Tree of Life (or any large tree) in a zoomable interface (Figure 1). This tool solves many issues that previous methods had, such as the possibility to display nodes with more than two descendants (multifurcations) and the possibility to explore smoothly very large trees of more than 2 million species.
Lifemap relies on tools that were developed for cartography, especially in the context of the OpenStreetMap project, and proposes a new way of representing hierarchical structures. The exploration of the ToL in Lifemap is similar to the exploration of a geographic map: by zooming and panning. It is also possible to click on every node and tip of the tree to have access to additional information (description, picture, link to external resources, etc.), and to search for “routes” in the tree of Life (Figure 1). Finally, an efficient search engine allows for an immediate retrieval of any node or tip by its Latin or its common name.
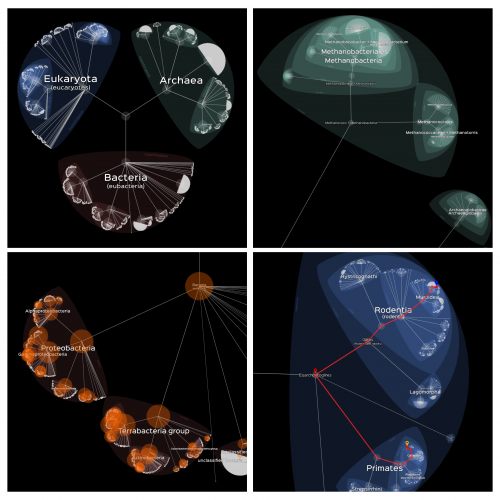
Lifemap comes in three versions, depending on the tree that is displayed and the information that is associated to nodes and tips when clicking. A first version displays the tree proposed by the Open Tree of Life (OTOL) project, a second one displays the whole NCBI taxonomy, and a third one displays a simplified version of the taxonomy with pictures and descriptions when clicking on the nodes (this last version is also available as an App for phones and tablets on the Android market).
In the near future, I would like the NCBI and the OTOL versions of Lifemap to be proper “hubs” giving access to a multitude of information of relevance to researchers in various fields. Because I personally work on comparative genomics, I started adding information on the number of genomes that are fully sequenced according to the NCBI database (Figure 1, bottom-left). This information can be displayed directly on the interactive tree. It is useful for easily identifying species, close to our species of interest, whose genome is available. I also added links on every node and tip to the corresponding web page on the NCBI web site and on other taxonomy web resources. These examples show how flexible Lifemap is at welcoming information from various sources.
I hope that developmental biology will be a field for which information could be displayed on Lifemap as well? I will be happy to consider such options if some interest is declared!
Useful links:
Lifemap welcome page: http://lifemap.univ-lyon1.fr
PLOS paper presenting Lifemap: http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.2001624
Lifemap on the Android Play Store: https://play.google.com/store/apps/details?id=fr.univ_lyon1.lifemapcom&hl=en
References:
Dobzhansky, T. (1973). Nothing in Biology Makes Sense Except in the Light of Evolution. American Biology Teacher, 35(3): 125-129.
de Vienne, DM. (2016). Lifemap: Exploring the Entire Tree of Life. PLOS Biology, 14(12): e2001624.
A short introduction to Lifemap:


 (9 votes)
(9 votes)
Dear Damien
Thank you so much for creating this valuable tool!
I’ve recently started teaching biology to 15-18 year-olds, and will be boarding the subject of taxonomy and the tree of life in a few weeks. I would like my pupils to learn independently about the tree of life by exploring your app. To guide them, I’m thinking of preparing a number of questions that they need to find the answer to, by looking through the map. Yet, maybe you already know of teachers’ resources that were created based on this app? This can also be in French.
Thank you in advance for your reaction!
Happy holidays,
Jiska
I am really fascinated by this project, quite similar to onezoom. The only thing I find unexpected is why you didn’t develop this tree by putting in one direction the time. I was hoping to come up with something that would look a bit like the tracks that slowly develop upon arrival at a giant terminal train station, with continuous branches and the possibility of zooming in, so that you arrive at the “station” with all the species existing today (t=now). It would, in my humble opinion, be clearer, but I understand that the technical difficulties might be difficult to solve. However, I wonder if you have thought about it.
Greetings and congratulations again!
Thanks you so much for this tool, it is an absolute lifesaver!