A mouse embryo atlas using HREM data
Posted by DMDD, on 23 May 2016
For the first time, a reference mouse embryo atlas has been created using HREM image data. For other embryo imaging methods such as micro-CT, a reference embryo atlas has previously been shown to be the basis of automated phenotyping (Wong et al, 2014). This new work (a collaboration between the DMDD programme and the Mouse Imaging Centre in Toronto) is a proof of principle for automated phenotyping of HREM data, which can now be tested in more detail.
COMPARING LIKE WITH LIKE
In order to phenotype an embryo, we need to compare it to another embryo that we consider to be normal. With histology this can be very difficult, as it’s virtually impossible to take exactly the same slice through two embryos.
So this leads to the question ‘have I really observed a phenotype, or am I simply looking at a different part of the embryo?’
The ideal scenario for a phenotyper is to have exactly equivalent slices of anatomy to compare. They can then do a direct visual comparison, or an algorithm can be used to do a statistical comparison, therefore automating the process to some degree.
A REFERENCE EMBRYO ATLAS
Using a technique previously employed with micro-CT data (Wong et al, 2012), the team have created a reference embryo atlas from HREM data. The atlas can be used to find these equivalent sections of anatomy.
21 wild-type embryo image stacks were merged together to give an unbiassed ‘average embryo’. Every possible pair of embryos was compared in terms of displacement. The embryos were then combined together so that the displacement for all embryos to join the atlas was a minimum. This technique avoids giving undue weighting to any one embryo in the average.
As HREM data is so large, the resolution was scaled back first to make the computation feasible.
For each individual embryo, a different deformation field is needed to bring the image into alignment with the average. The inverse of this field can then be used to propagate back from any plane in the reference embryo to a section of the original input embryo.
This means it’s possible to find equivalent embryo sections to compare, ensuring that any differences observed are much more likely to be genuine phenotypes.

It’s important to note that these sections are never planes but crinkly slices through the embryo. This means that the sections couldn’t have been identified by manually scrolling through the image stacks.
AUTOMATED PHENOTYPING
Automated selection of equivalent embryo sections is the starting point for automated phenotyping. Given the time-consuming nature of phenotyping embryos, and the large task of phenotyping many mouse lines, automated phenotyping is an attractive prospect to many. It can act as a guide – a primary screen to highlight areas of interest to manual phenotypers.
The next step, then, is to compare other HREM embryo data with this reference atlas.
The results of automated primary screens are typically presented as a heatmap (Wong et al, 2014), highlighting areas of interest. The authors showed a proof of principle for an automated primary screen of HREM data by separating the embryos into male and female groups. After selecting equivalent sections through the gonads to compare between the two groups, they were able to automatically distinguish statistically significant differences in this region.
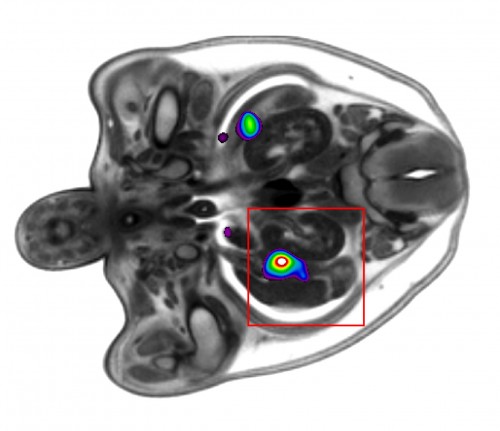
This is an exciting result for the future of automated phenotyping of HREM mouse embryo data, which will be tested much more extensively over the coming months.
This post originally appeared on the DMDD blog, Annotations (blog.dmdd.org.uk).
DMDD is a systematic study of embryonic-lethal mouse gene knockouts, and provides a database of high-resolution image and phenotype data – a valuable resource for both developmental biologists and clinicians. Visit the website to find out more: dmdd.org.uk
READ THE FULL PAPER
R. Mark Henkelman1, Miriam Friedel1, Jason P. Lerch1, Robert Wilson2, Tim Mohun2 (2016)
Comparing homologous microscopic sections from multiple embryos using HREM
Developmental Biology [Epub ahead of print], DOI: 10.1016/j.ydbio.2016.05.011
1 Mouse Imaging Centre (MICe), Hospital for Sick Children, University of Toronto, Toronto, Canada
2 The Francis Crick Institute Mill Hill Laboratory, London, UK
REFERENCES
M. D. Wong1, Y. Maezawa2, J. P. Lerch1, R. M. Henkelman1 (2014)
Automated pipeline for anatomical phenotyping of mouse embryos using micro-CT
Development, DOI: 10.1242/dev.107722
1 Mouse Imaging Centre (MICe), Hospital for Sick Children, University of Toronto, Toronto, Canada
2 The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Canada
M. D. Wong1,2, A. E. Dorr1,3, J. R. Walls4, J. P. Lerch1,2, R. M. Henkelman1,2 (2012)
A novel 3D mouse embryo atlas based on micro-CT
Development, DOI: 10.1242/dev.082016
1 Department of Medical Biophysics, University of Toronto, Toronto, Canada
2 Mouse Imaging Centre (MICe), Hospital for Sick Children, University of Toronto, Toronto, Canada
3 Sunnybrook Hospital, Toronto, Canada
4 Regeneron Pharmaceutecals, Tarrytown, USA


 (No Ratings Yet)
(No Ratings Yet)