The people behind the papers: Ehsan Pourkarimi & Iestyn Whitehouse
Posted by the Node Interviews, on 25 January 2017
How does a developing embryo coordinate DNA replication with gene transcription? This fundamental question is the focus of today’s paper, recently published in eLife. We caught up with lead author Ehsan Pourkarimi and his supervisor Iestyn Whitehouse, Assistant Professor in the Sloan Kettering Institute’s Molecular Biology programme, to hear the story behind the work.

Iestyn, can you tell us your scientific biography and the general focus of the Whitehouse lab?
IW My first project as a graduate student was using biochemistry to study how nucleosomes are repositioned along DNA by so-called chromatin remodelling complexes in the lab of Tom Owen-Hughes in Dundee, Scotland. After that, I wanted to learn more about chromatin in vivo so I joined Toshi Tsukiyama’s lab at the Fred Hutch in Seattle. There, I learned to use budding yeast and genomics and studied how genes are activated and repressed by nucleosomes.
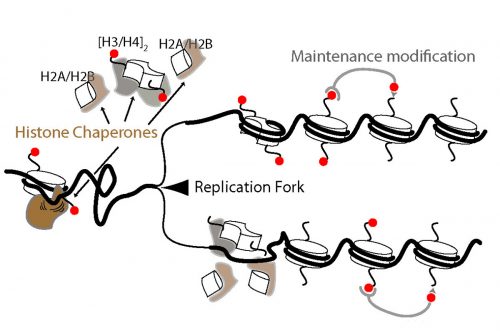
I’ve always been interested in the question of whether chromatin states can be inherited through generations. When I started my own lab, I wanted to focus on understanding the basics of DNA replication in vivo with the hope that would allow a better understanding of how chromatin may be intertwined with DNA replication. More generally, my lab works on chromatin and how it relates to gene expression and DNA replication.
Sloan-Kettering would seem an ideal place for a molecular biologist dabbling in developmental biology?
IW Absolutely, although I must admit I was first attracted to come here because of the strength of the research in DNA replication and genomic integrity. With my background in chromatin and gene transcription I wanted to be around people who could help me with some project ideas I had about DNA replication. As the projects in my lab have developed it’s a tremendous benefit to be able to interact with developmental biologists here at Sloan Kettering.
And Ehsan, how did you come to work in New York with Iestyn?
EP During both my undergraduate and PhD research I mostly focused on how cells commit suicide or more precisely apoptosis, a genetically regulated process of programmed cell death. I did my PhD in Anton Gartner’s lab in Scotland, for a few exciting years that turned out to be scientifically very productive. Toward the end of my PhD, I became fascinated about the field of chromatin biology and epigenetics. The last year of my research in Scotland mostly revolved around understanding how cells respond to acute stress conditions such as starvation. Further, I was interested in a possible role of chromatin remodelers and histone modifications in such stress responses. This research led to the discovery of the function of histone H3.3 variants in both starvation response and aging.
It was around the same time that Iestyn and Duncan Smith (a former postdoc in Iestyn’s lab) for the first time purified and sequenced Okazaki Fragments in budding yeast; the biased distribution of yeast Okazaki fragments perfectly matched the previously known replication origins. Right then I was sure that this approach could be utilized in metazoans. I was motivated to find out if we finally could get a complete picture of metazoan replication origins. So I decided to shift my research from how cells die to how they are born. Iestyn’s lab was the ideal place for my new interest. After meeting with Iestyn and discussing the idea of establishing C.elegans system in the lab, I moved to New York. Iestyn is among the top scientists in the field of chromatin biology and open to new ideas and approaches, which has made my transition to this field easy and exciting. Three years later, I can confidently tell you that moving to New York and switching my research field was one of my best decisions.
“I was motivated to find out if we finally could get a complete picture of metazoan replication origins. So I decided to shift my research from how cells die to how they are born”
Your previous work has dealt with chromatin and replication in S. cerevisiae. What made you decide to use C. elegans in this paper?
IW Although I enjoy working with yeast, I think I’d work with any organism if there were an interesting question we could address. In terms of this project, I was interested in testing whether our general findings about the relationship between chromatin and DNA replication from budding yeast would hold up in another organisms. Ehsan came to me with many years of experience working with C. elegans; he told me he wanted to map DNA replication in worms – I said go for it!
Was it difficult to apply techniques originally used in yeast to developing worms?
IW & EP If you would ask me this question 3 years ago I would bet against myself! Having said that, the beauty of having questions is that they force you to challenge yourself. I wouldn’t necessarily say it was difficult, but it required hard work. We needed tons of optimization and we went through many many trails and errors. The beauty of C. elegans is its ease of genetic manipulation. Once we knew how strongly we needed to knock down lig-1, we were able to radiolabel and visualize Okazaki fragments. The most challenging part was purifying Okazaki fragments without causing too much DNA damage to developing embryos.
“The beauty of having questions is that they force you to challenge yourself. I wouldn’t necessarily say it was difficult, but it required hard work”
Can you give us the key results of your paper?
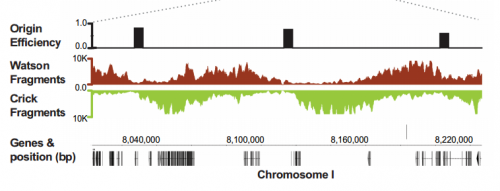
IW & EP We have studied DNA replication in multicellular Caenorhabditis elegans embryos and have identified a distinct set of replication origins. Interestingly, replication origins are marked by histone proteins containing specific modifications such as H3K27ac. These modifications are typically associated with sites of active gene transcription. This finding raises a key question: Does transcription define the sites of replication origins, or do replication origins define where transcription will occur?
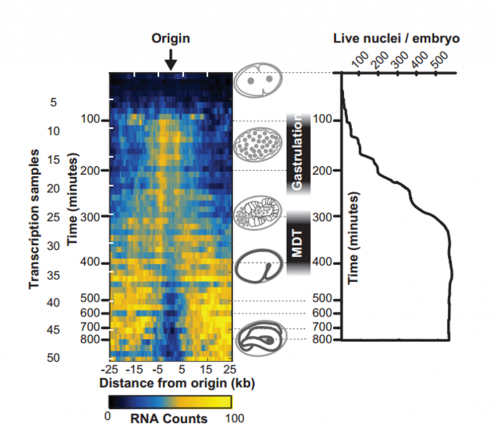
To answer this, we identified replication origins in very early embryos (pre-gastrula); at a stage in embryogenesis in which most genes are inactive. Surprisingly, we find that replication origins are present at this very early stage, which shows that replication origins precede active transcription. Nevertheless, we found that once transcription begins, it does so in close proximity to the pre-defined origins. This close association of gene transcription and DNA replication persists until the embryonic cells stop dividing and experience a profound change in their gene activity and morphology. At this stage of development, the association between DNA replication and gene transcription breaks down and genes located farther away from replication origins begin to be transcribed. Our work suggests that the genome has evolved to couple DNA replication with gene transcription in rapidly dividing cells.
Does this work imply that the layout of genome is tied to the mode of development of the animal?
IW & EP Yes, it’s an attractive possibility. The rapidity of S phase in embryonic cells – which, in C. elegans is on the order of 15-20 minutes – is probably a strong determinant of genome organization. To replicate a 100mb genome in that timeframe you need a replication origin about every ~75kb. Like other organisms, replication origins in C. elegans are associated with transcriptionally active chromatin marks, so we think that the requirement for regularly spaced origins has influenced gene organization: genes associated with growth are clustered near replication origins. We’re very interested in the idea that replication origins may be intimately tied to nearby gene activity. One consequence of this is that activation and inactivation of origins will profoundly alter not only the transcription program but also the kinetics of S phase.
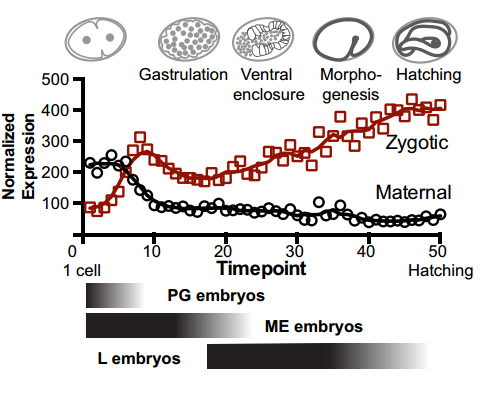
And your work might also have implications for the proposed ‘mid-developmental transition’ across phyla?
IW/EP One of the striking features of the MDT is the switch from the expression of “growth” to “differentiation” genes. Broadly speaking you want to express growth genes in cycling cells (when you’re replicating DNA) and differentiation genes when you’re not. If you couple the expression of growth genes with replication origin function then inactivation of replication origins may be part of the trigger to shift the transcriptional program from growth to differentiation. Of course, this is mostly speculation, nature is full of surprises and we’ll wait and see how it turns out.
You reference and even include comparison data from quite a few papers published in the seventies and eighties. What was it like to revisit this old work?
IW & EP It’s always rewarding to go back and dig through the older literature – we cite some classic papers that have stood the test of time. It’s also very important understand what current dogma is based on: all too often what we think we know isn’t really supported by the facts and when you go back to the literature you realize that there are large gaps in our knowledge. In this paper we’ve found that replication origins are established very early in development, which goes against the dogma; the old data isn’t wrong, rather we’ve found that C. elegans behaves differently to Xenopus and Drosophila.
“All too often what we think we know isn’t really supported by the facts and when you go back to the literature you realize that there are large gaps in our knowledge”
When doing the research, did you have any particular result or eureka moment that has stuck with you?
EP The very first time I experienced such feeling was during my undergraduate studies in Budapest in the lab of Krisztina Takacs. At the time, I was working on the C. elegans counterpart of the anti-metastasis gene nm23. After long hours of staring into a microscope, I could clearly see nm23 is modulating the RAS/MAPK pathway; a very unexpected finding that made me lose sleep for many nights. Back then, in my innocence, I thought I would never forget the joy and the satisfaction I felt in that moment. Years later in a meeting one of my former professors congratulated me on passing the test of time on the same finding, and I realized that that was my eureka moment. As for this research, I will have my eureka moment once time puts its stamp of approval on our findings.
And what about your plans following this work?
EP We have opened up multiple interesting questions in this field. Currently we are interested in looking at replicating pattern in highly specialized somatic cells, which are transcriptionally dissimilar to embryonic cells. We know that histone modifications are altered in such cells, which indicate a possible variant replication profile. At the same time, we are using different genetic screens to pinpoint upstream component(s) of replication initiation.
Do you plan to continue this developmental strand in the Whitehouse lab?
IW Yes – although I wouldn’t classify myself as a developmental biologist just yet. Maybe the best way to describe my thinking is that I’ve come to realize (rather belatedly) that context is all-important. As molecular biologists we typically study processes and systems far removed form the context in which they typically function. Mostly, this is dictated by practicality of getting the experiments done. But the cost of the practicality is often paid by not being able to contextualise the results. In the case of chromatin, for example, I think we understand the basic principles of what chromatin is, but there are many aspects – not least the biological functions of histone modifications – that are still a real mystery. Generally speaking, I think we’ll figure out some of the mysteries if we can identify the biological context in which certain chromatin states and histone modifications come in to play. This is where developmental biology will be especially powerful – we’ll see how far we get!
“As molecular biologists we typically study processes and systems far removed form the context in which they typically function…the cost of the practicality is often paid by not being able to contextualise the results”
Ehsan Pourkarimi, James M Bellush & Iestyn Whitehouse (2016) Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. eLife 5:e21728
Browse the People Behind the Papers archive here.


 (1 votes)
(1 votes)