A Day in the Life of a Turtle lab
Posted by Juan Pascual-Anaya, on 9 December 2014
We are Juan Pascual-Anaya and Tatsuya Hirasawa, two postdoctoral fellows at RIKEN (Kobe, Japan), in the laboratory of Shigeru Kuratani. We both have been working for the last 4-5 years using turtles as a model organism in the laboratory, studying different aspects of the carapace development.
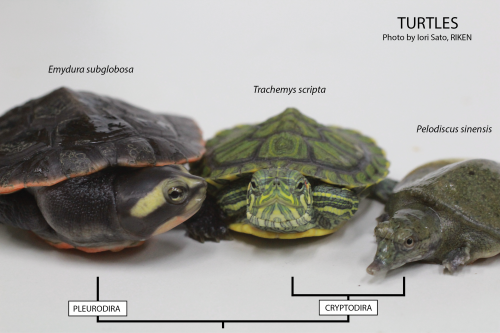
Three different turtles used in our lab.
The turtle shell, composed of dorsal and ventral moieties, the so-called carapace and plastron, respectively, is an outstanding example of what in evolutionary biology is called a morphological novelty. If you think of the body plan of a general tetrapod, you’ll realize that the turtle is the only one with such a display of the ribs in the dorsal part of the animal, disposition in which during development the shoulder girdle eventually ends up below the ribs (when generally it is outside the ribcage –we know everyone would be touching his or her shoulder blade at this moment–). The turtle shell is a genuine evolutionary innovation, as it has no counterpart in other animals. We are interested in uncovering the developmental mechanisms underlying this morphological novelty.
Using the turtle as a model in the laboratory is not so different from using for example the chicken, but with one big disadvantage: turtles only lay eggs seasonally, meaning that we need to concentrate all of our efforts to perform in vivo experiments during the summer. We mostly work with the Chinese soft-shell turtle, Pelodiscus sinensis (picture above, on the right). And why is that? Well, the Chinese soft-shell turtle is a delicacy found in the Asian market. Here in Japan, where it is called suppon (スッポン), it is used for the preparation of an exquisite dish, the suppon-nabe in which the turtle is cooked in a soup with rice. This fact makes it possible to directly order eggs to a local farm. And that is exactly what we usually do. During the reproductive season, we order around 100 eggs weekly. Then, depending on the availability, we’ll get that or less… or nothing. For example, when the weather worsens, the turtles do not lay eggs.
This is what we usually do in a day, or more strictly, along the whole season:
When the eggs arrive, they do it slightly burrowed in a wet sand or mud, with no so much water, just enough to keep the humidity within the box. We then need to dig them out, clean the excess of sand and put them on a clean and wet litter. We then culture them at 30 degrees. Fortunately, the development of P. sinensis is quite synchronous, more than in chicken, and we can know exactly in which stage the embryos will be everyday. There are several staging tables available for many turtles. For P. sinensis, our supervisor Shigeru Kuratani reported such standard table back in 2001 (Tokita and Kuratani, 2001).
Eggs of Pelodiscus sinensis just after arriving to the lab.
So, we then will use most of the eggs to get embryos at given stages in which we would like to study the expression pattern of a gene, that is to do in situ hybridizations. The eggs of P. sinensis are quite small, so we need to manage them with care to avoid just smashing them and losing embryos. We use fine forceps and dissecting scissors to open them, and if the embryo is old enough, we peel the egg’s shell little by little, until the embryo is fully visible. When the embryos are way too young, for instance until stage 11 or so (like stage HH16 of chicken), the embryo can be dragged completely in a piece of egg’s shell, put this upside-down, and then extracted from the egg shell. It’s like using a holed piece of paper to extract young embryos of chicken, but using the shell itself. Then, the embryos are just fixed in paraformaldehyde as in standard protocols.
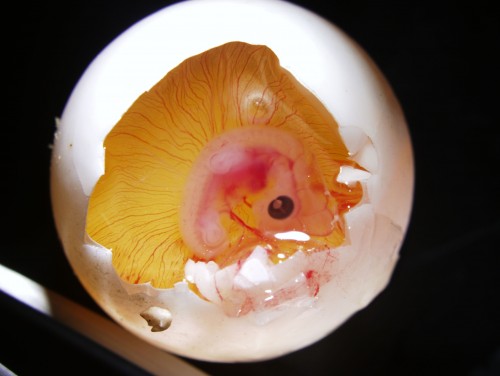
The eggs, on the other hand, can be windowed as the chicken ones, but with a very small hole. Then, we can assay the embryos with a number of techniques. This includes electroporation, DiI labellings, embryological operations, and so on. In one experiment that I have been involved recently, I was introducing morphogen-soaked beads into the lateral trunk of the turtle embryo. This is just as it has been done in chicken, but in miniaturized egg! You can see in the picture below a windowed egg and the embryo inside.
Windowed egg of P. sinensis showing an embryo at stage TK13-14
Last, we sometimes need just hatched juveniles. P. sinensis juveniles usually hatch around a month and a half after they arrive to the lab, and here you can see the birth moment in the following video:
I hope you liked what can be done using turtles in the lab.
Thanks!
Juan and Tatsuya.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.



 (15 votes)
(15 votes)
I am from chicken lab, I liked this article very much. I am quite happy to know and learn from this article that there are pretty much similar things we do with chicken eggs.
Good one.
Thanks, Sksindhu! I have been working on chicken embryos (chicken-quail chimera experiments, etc.) in another research project, too.
Great piece! It’s great to know about turtles as I find them amusing animals and didn’t know much about their development, or labs that work to learn more about this. I was wondering if their genome has been sequenced, and what kind of tools you have available but maybe that’s good material for a future post?…
Good luck with experiments, enjoy the busy summers!
Thanks for the comment! Yes, there are actually three genomes available, including the one of P. sinensis. We published last year that one together with the green sea turtle’s, Chelonia mydas (http://www.nature.com/ng/journal/v45/n6/full/ng.2615.html), and at the same time the group of Bradley Shaffer published the western painted turtle genome, Chrysemys picta bellii (http://genomebiology.com/2013/14/3/R28).
About tools, if you talk about genomics, we are on that. For instance, I am currently analysing some RNA-seq and ChIP-seq data related with the carapace formation.
This is really amazing that you guys can also do CHIP-Seq on turtle samples. Is it easy to get hold of antibodies that work well for turtle especially those that may work in CHIP?
Hi WYE. Thanks for your comment!
I am analysing histone modifciation ChIP-seq data, and for that I used commercially available antibodies, cited elsewhere, and they have been successfully used in a wide range of organisms, from Drosophila to human stem cells. Histones are extremely conserved, and thus antibodies against them usually work pretty well in distantly related models.