In Development this week (Vol. 139, Issue 10)
Posted by Seema Grewal, on 23 April 2012
Here are the highlights from the current issue of Development:
A TOR de force in the haematopoietic niche
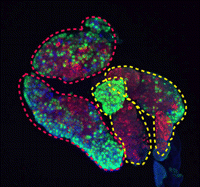 During development and homeostasis, it is essential to coordinate growth with the availability of nutrients. The interconnected insulin/IGF (IIS) and target of rapamycin (TOR) pathways integrate tissue growth with dietary conditions in Drosophila, and now Marc Haenlin and co-workers show that these pathways play a crucial role during haematopoiesis in the Drosophila lymph gland (p. 1713). The larval lymph gland contains a group of stem-like progenitor blood cells (prohaemocytes) that are kept in an undifferentiated state by cells of the posterior signalling centre (PSC), which serves as the stem cell niche. The researchers show that the IIS and TOR pathways regulate the size of the haematopoietic niche by regulating cell size and cell proliferation in the PSC. In addition, they show that IIS and TOR signalling are required in prohaemocytes to control their maintenance, and disruption of these pathways, induced genetically or by starvation, results in the precocious differentiation of these progenitors. Importantly, these studies highlight that blood cell development is coupled with nutritional status.
During development and homeostasis, it is essential to coordinate growth with the availability of nutrients. The interconnected insulin/IGF (IIS) and target of rapamycin (TOR) pathways integrate tissue growth with dietary conditions in Drosophila, and now Marc Haenlin and co-workers show that these pathways play a crucial role during haematopoiesis in the Drosophila lymph gland (p. 1713). The larval lymph gland contains a group of stem-like progenitor blood cells (prohaemocytes) that are kept in an undifferentiated state by cells of the posterior signalling centre (PSC), which serves as the stem cell niche. The researchers show that the IIS and TOR pathways regulate the size of the haematopoietic niche by regulating cell size and cell proliferation in the PSC. In addition, they show that IIS and TOR signalling are required in prohaemocytes to control their maintenance, and disruption of these pathways, induced genetically or by starvation, results in the precocious differentiation of these progenitors. Importantly, these studies highlight that blood cell development is coupled with nutritional status.
A MAP(K) of germline self-renewal
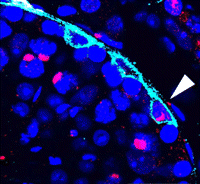 Spermatogonial stem cells (SSCs) have the remarkable ability to self-renew and support spermatogenesis throughout life. It is known that fibroblast growth factor 2 (FGF2) promotes SSC self-renewal but the factors acting downstream of FGF2 are unknown. Here, Takashi Shinohara and colleagues show that FGF2 regulates SSC self-renewal via MAP2K1 and the Etv5 and Bcl6b genes (p. 1734). Using an in vitro mouse germline stem (GS) cell culture system, the authors show that GS cells require FGF2 for continuous proliferation, and that a specific MAP2K1 inhibitor reduces GS cell proliferation and MAP2K1 phosphorylation. By analysing target genes that are regulated by MAP2K1, the researchers identify Etv5 and Bcl6b, and show that overexpression of these genes in GS cells promotes proliferation in an FGF2-independent manner, confirming that they act downstream of MAP2K1. Furthermore, transplantation of Bcl6b-expressing GS cells into mouse testes induces germ cell tumour formation, suggesting that excessive self-renewal can promote tumourigenesis. The identification of these genes provides key insights into the mechanisms controlling SSC self-renewal.
Spermatogonial stem cells (SSCs) have the remarkable ability to self-renew and support spermatogenesis throughout life. It is known that fibroblast growth factor 2 (FGF2) promotes SSC self-renewal but the factors acting downstream of FGF2 are unknown. Here, Takashi Shinohara and colleagues show that FGF2 regulates SSC self-renewal via MAP2K1 and the Etv5 and Bcl6b genes (p. 1734). Using an in vitro mouse germline stem (GS) cell culture system, the authors show that GS cells require FGF2 for continuous proliferation, and that a specific MAP2K1 inhibitor reduces GS cell proliferation and MAP2K1 phosphorylation. By analysing target genes that are regulated by MAP2K1, the researchers identify Etv5 and Bcl6b, and show that overexpression of these genes in GS cells promotes proliferation in an FGF2-independent manner, confirming that they act downstream of MAP2K1. Furthermore, transplantation of Bcl6b-expressing GS cells into mouse testes induces germ cell tumour formation, suggesting that excessive self-renewal can promote tumourigenesis. The identification of these genes provides key insights into the mechanisms controlling SSC self-renewal.
Notch tips the balance in the pancreas
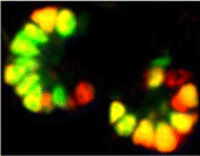 In the developing pancreas, the branched epithelium can be separated into tip and trunk regions, with the tip domain generating acinar cells, and the trunk domain differentiating to endocrine and duct fates. Although Notch signalling is known to be important for proper pancreatic development, particularly in maintaining the progenitor state and inhibiting premature endocrine differentiation, its precise roles in regulating cell fate remain unclear. Here (p. 1744), Jan Jensen and co-workers disrupt Notch signalling in the mouse in a mosaic fashion, revealing a function for Notch in regulating trunk versus tip cell fate. Overexpression of a dominant-negative Mastermind protein, which blocks Notch-dependent transcription, leads to loss of endocrine and duct cells, suggesting that Notch signalling promotes trunk cell identity. Mechanistically, Notch promotes the expression of the trunk-specific transcription factor Nkx6.1, via direct binding of RBP-jκ at the Nkx6.1 promoter. These data thus establish a crucial role for the Notch pathway in directing endocrine and duct cell differentiation in the pancreas.
In the developing pancreas, the branched epithelium can be separated into tip and trunk regions, with the tip domain generating acinar cells, and the trunk domain differentiating to endocrine and duct fates. Although Notch signalling is known to be important for proper pancreatic development, particularly in maintaining the progenitor state and inhibiting premature endocrine differentiation, its precise roles in regulating cell fate remain unclear. Here (p. 1744), Jan Jensen and co-workers disrupt Notch signalling in the mouse in a mosaic fashion, revealing a function for Notch in regulating trunk versus tip cell fate. Overexpression of a dominant-negative Mastermind protein, which blocks Notch-dependent transcription, leads to loss of endocrine and duct cells, suggesting that Notch signalling promotes trunk cell identity. Mechanistically, Notch promotes the expression of the trunk-specific transcription factor Nkx6.1, via direct binding of RBP-jκ at the Nkx6.1 promoter. These data thus establish a crucial role for the Notch pathway in directing endocrine and duct cell differentiation in the pancreas.
Eve and Grain guide the way for axon pathfinding
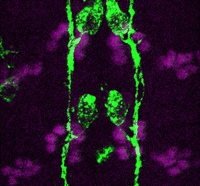 Accurate axonal pathfinding relies on the tightly regulated expression of guidance cues and their receptors, but the links between transcriptional regulators and downstream guidance factors are poorly understood. Genetically amenable Drosophila motoneurons provide an ideal system for analysing the control of guidance receptor expression. It is known that two transcription factors, Even-skipped (Eve) and Grain (Grn) are expressed in the aCC and RP2 motoneurons, and that projection of these neurons to the muscle requires the Netrin receptor Unc-5. Now, Juan-Pablo Labrador and colleagues dissect out the relationships between these factors (p. 1798). The researchers find that Eve and Grn independently promote Unc-5 transcription, and that both are required to generate sufficient Unc-5 expression for proper pathfinding – likely via an enhancer element in unc-5 intron 5. Overexpression of both Eve and Grn in another motoneuron population induces ectopic Unc-5 and hence axonal redirection. Thus, the combinatorial effects of these two transcription factors together direct expression of the key guidance receptor, and so define the axon’s path.
Accurate axonal pathfinding relies on the tightly regulated expression of guidance cues and their receptors, but the links between transcriptional regulators and downstream guidance factors are poorly understood. Genetically amenable Drosophila motoneurons provide an ideal system for analysing the control of guidance receptor expression. It is known that two transcription factors, Even-skipped (Eve) and Grain (Grn) are expressed in the aCC and RP2 motoneurons, and that projection of these neurons to the muscle requires the Netrin receptor Unc-5. Now, Juan-Pablo Labrador and colleagues dissect out the relationships between these factors (p. 1798). The researchers find that Eve and Grn independently promote Unc-5 transcription, and that both are required to generate sufficient Unc-5 expression for proper pathfinding – likely via an enhancer element in unc-5 intron 5. Overexpression of both Eve and Grn in another motoneuron population induces ectopic Unc-5 and hence axonal redirection. Thus, the combinatorial effects of these two transcription factors together direct expression of the key guidance receptor, and so define the axon’s path.
Planar cell polarity: fattened up
 The atypical cadherin Fat (Ft) is crucial for planar cell polarity (PCP) in Drosophila. Four ft homologs (Fat1 to Fat4) have been identified in mammals, but the functional roles of these homologs and any possible redundancies between them are unclear. Here, Helen McNeill and colleagues study the genetic interactions between mammalian Fat genes and show that Fat proteins act both synergistically and antagonistically to regulate multiple aspects of tissue morphogenesis in mice (p. 1806). For example, the authors show that Fat1 and Fat4 synergise during kidney, cochlea and cranial neural tube morphogenesis. Importantly, the researchers also show that the effects of Fat4 are modulated by atrophins, which are known components of PCP signalling in Drosophila, suggesting that Fat-atrophin interactions play an essential and conserved role in planar polarity. These findings reveal a high degree of complexity in mammalian PCP and highlight the wide-ranging effects of Fat cadherins on animal development.
The atypical cadherin Fat (Ft) is crucial for planar cell polarity (PCP) in Drosophila. Four ft homologs (Fat1 to Fat4) have been identified in mammals, but the functional roles of these homologs and any possible redundancies between them are unclear. Here, Helen McNeill and colleagues study the genetic interactions between mammalian Fat genes and show that Fat proteins act both synergistically and antagonistically to regulate multiple aspects of tissue morphogenesis in mice (p. 1806). For example, the authors show that Fat1 and Fat4 synergise during kidney, cochlea and cranial neural tube morphogenesis. Importantly, the researchers also show that the effects of Fat4 are modulated by atrophins, which are known components of PCP signalling in Drosophila, suggesting that Fat-atrophin interactions play an essential and conserved role in planar polarity. These findings reveal a high degree of complexity in mammalian PCP and highlight the wide-ranging effects of Fat cadherins on animal development.
Complexity in the kidney
 The kidney comprises multiple cell types of both epithelial and mesenchymal origin, with highly defined regional subdivisions in the ductal systems. A full understanding of kidney development requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their results not only identify novel markers, but also reveal an unexpected degree of restriction in expression of many factors, suggesting that anatomically defined compartments may be further subdivided at the molecular level. Moreover, this in situ dataset provides a starting point to understand the transcriptional networks underlying cell type specification. As proof of principle, the authors use published microarray and expression data to bioinformatically identify putative targets of five transcription factors and to uncover potential network topologies. This valuable resource has been made available to the community via the GUDMAP database.
The kidney comprises multiple cell types of both epithelial and mesenchymal origin, with highly defined regional subdivisions in the ductal systems. A full understanding of kidney development requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their results not only identify novel markers, but also reveal an unexpected degree of restriction in expression of many factors, suggesting that anatomically defined compartments may be further subdivided at the molecular level. Moreover, this in situ dataset provides a starting point to understand the transcriptional networks underlying cell type specification. As proof of principle, the authors use published microarray and expression data to bioinformatically identify putative targets of five transcription factors and to uncover potential network topologies. This valuable resource has been made available to the community via the GUDMAP database.


 (No Ratings Yet)
(No Ratings Yet)