Off and On: it’s more complicated than we thought.
Posted by stevegis, on 23 January 2020
We learn fairly early on when becoming biologists that both development and an organism’s response to environmental stressors require turning the right set of genes on in the right cells, at the right time. Clearly, for “on” to be meaningful, other genes have to be “off,” and many disordered conditions are associated with misexpression of genes, from the classic Antennapedia fly to oncogenes in cancer. It’s natural to think of “off” as the default state, but we know it’s more complicated than that. For one thing, transcription factors, the proteins that orchestrate the transcriptional program in a given cell state, are highly pleiotropic. Pax6 in vertebrates is expressed in the developing eye and pancreas, and is necessary for specific gene expression in both contexts, and yet digestive enzymes are not secreted into the lens. There is another layer of regulation that can turn genes off, specifically, in cellular contexts where they are not supposed to be expressed.
In the decades that we (as a community) have been studying gene regulation, we have identified and dissected sequences that act as enhancers, turning on nearby genes in some element of their correct expression pattern, and others that act as silencers, turning them off. There have even been a handful identified, in eukaryotes as distantly related as plants and people, that have both activities, in different contexts. Enhancer studies are a pretty mature field: reporter assays, in which a piece of DNA is cloned near an exogenous gene under the control of a minimal promoter and any resulting expression examined in vivo, have been a workhorse of the field for decades. In Drosophila, my own model organism of choice, a single overnight collection of embryos from such a reporter strain can reveal the entire spatiotemporal pattern of activity of an enhancer across all stages of embryogenesis. This method has enabled the characterization of many thousands of Drosphila enhancers, and been extended to create Massively Parallel Reporter Assays (MPRAs), in which enhancer activity can be quantitatively measured up to genome-wide, at the expense of pattern information. Moreover, the combination of this wealth of enhancer information with the introduction of chromatin immunoprecipitation (ChIP)-based methods for assaying genomic occupancy of histone marks and regulatory proteins has enabled us to predict enhancers with reasonable accuracy, when a sufficiently large sample of the relevant cell type is available for ChIP experiments.
By contrast, the study of silencers has lagged. I believe that this is in large part just because it’s more technically challenging. The whole-embryo enhancer-reporter assay, for all that it can be fiddly and time-consuming, is in principle quite simple: put the DNA into flies, see where the reporter gene is expressed. If you want to see silencing, you have to test the candidate silencer in the context of an element that would otherwise drive expression. There are for instance beautiful classic studies1 in the fly embryo that used a silencer that acts in a stripe and an enhancer that acts in an orthogonal stripe, but this requires having some idea about the activity pattern of the silencer before you start making your flies. Or, as in MPRAs for enhancers, you can look at an enhancer that acts in a single cell type, and then look genome-wide. But there is a problem: other promoters in your reporter construct can compete for the attention of an enhancer and reduce its activity on the reporter promoter. While interesting, this phenomenon is not silencing per se, and it creates artifacts in a silencer-reporter assay that can swamp your genuine signal, as we rediscovered to our chagrin. For all these reasons, silencers represent a relatively juicy target for discovery and characterization.
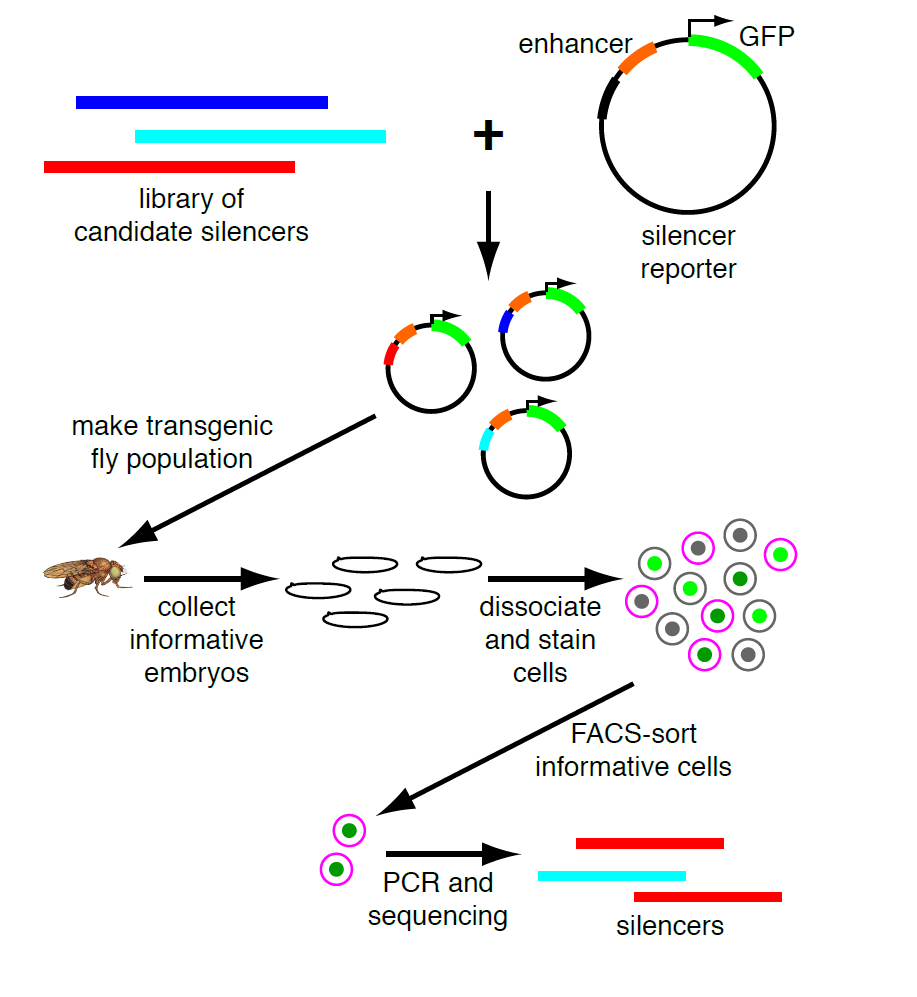
My PI, Martha Bulyk, likes to think of our research program as a virtuous cycle, where biological questions motivate technology development and new technologies open up new questions. So when we published a paper describing a moderately parallel reporter assay for in vivo use in defined cell populations derived from developing embryos2, and coincidentally found a VERY strong, nearly ubiquitous enhancer, she realized that we could adapt this to look for silencers. Our method is based on fluorescence-activated cell sorting with a GFP reporter, and this enhancer is strong enough that cells that got the transgene are completely separate from those that didn’t. Thus we can sort the cells that fall in between these two populations, and these are the ones in which GFP expression is active but silenced. Sorting on a separate marker gives us cell-type specificity; PCR recovery of the inserts and high-throughput sequencing are the readout. (see Figure)
The throughput of this method is limited by the fly-transformation step to hundreds of sequences tested in parallel, so it’s not suitable for a genome-wide experiment. We had a few specific hypotheses about potential silencers and questions we wanted to test. One of the most reliable sources of enhancer predictions for in vivo testing has been ChIP for the coactivator proteins that transcription factors work by recruiting, particularly the p300/CBP histone acetyltransferases. So we thought the best possible source of silencers would be ChIP data for corepressors, and some of these datasets were already available in the fly embryo. I was particularly interested in comparing binding sites for the Groucho and CtBP corepressors, because they had canonically been (tentatively) associated with long-range and short-range repression, respectively. Short-range repressors had been characterized as acting within an enhancer to limit its activity, and careful studies of designed mutations of individual elements (such as 3) had shown that even moving a short-range repressor’s binding site from 50 bases away to 150 bases away from an enhancer would eliminate its activity. Since our cloning strategy pads all tested elements and then adds an additional 100 bases between them, we thought that short-range repressors would not be active in our assay. Long-range repressors, by contrast, act on distant promoters, even when far away from otherwise active enhancers. For these reasons, we hypothesized that Groucho-occupied sites would be a much richer source of silencers than CtBP-occupied sites.
Another good source of enhancer predictions has been ChIP for the modified histones that define an “enhancer chromatin state,” first observed as enrichment in the wealth of known enhancers and then confirmed by testing of additional elements so predicted. But there is no similarly defined “silencer state” yet known. There are histone marks (H3K9me3 and H3K27me3 in particular) that are associated with inactive chromatin, but conceptually this can represent the end product of silencer activity, i.e. its target, and not necessarily the region that contains the regulatory information for silencing. We hypothesized that where marks of active regulatory regions (like H3K4me1 or DNase accessibility) coincided with repressive marks, this might be another source of candidate silencers, so we tested some of these regions mined from published genome-wide data.
Finally, several of the silencers that have been studied and reported previously were sequence elements that could silence in one cell type but were enhancers in another. I myself was most interested in the question of how widespread a phenomenon this would prove to be: how often is a silencer also an enhancer, and conversely how often is an enhancer also a silencer? Since we were looking for silencer activity in the embryonic mesoderm, we could use the tremendous genetic resources available in flies—in this case, the REDfly database of published enhancers and the Berkeley Drosophila Genome Project database of gene expression patterns—to identify hundreds of enhancers for genes that are not expressed in the mesoderm.
As I foreshadowed a few paragraphs ago, we neglected to account for promoter competition when designing our library. Like any decent experimentalists, we included controls, and some of our negative controls were enhancers that drive widespread mesodermal expression, since we reasoned that they cannot also be mesodermal silencers at the same time. Yet some of these scored positive in our assay, and these turned out to overlap promoters. In fact, promoter overlap was the most enriched feature in our first set of positives. Promoter competition undoubtedly has a role in establishing the regulatory logic of the genome, but it’s not the same as the silencing activity we were looking for, and the ability of these elements to inhibit reporter gene expression in an integrated plasmid does not reflect their ability to silence their endogenous target genes. In subsequent experiments, we simply excluded promoters from our library of tested elements.
At last we could address the questions we designed our libraries to ask4, and (unusually, in my experience) the main answer was pretty clear: silencers are enhancers. Like, pretty much all of them. The only class of input sequences enriched for mesodermal silencers was non-mesodermal enhancers. A few were also enhancers active in just a small subset of the mesoderm, and thus presumably silencers in one or more other mesodermal cell types. Very few of the corepressor binding sites scored positive in our assay—just three CtBP peaks and only one Groucho peak, which certainly doesn’t support the short range / long range distinction we had expected to see—and three of those four elements were embryonic enhancers when we tested them for that activity. You can’t prove a negative this way, of course. We did not, after all, test all the DNA. But we went looking for a class of silencers that are not also enhancers and, with one possible exception, we didn’t find them. What’s more, over 10% of all the enhancers we tested were also silencers. Now, I want to emphasize again that this bifunctionality was a known phenomenon. We did not discover this. But I wanted to find out how common a phenomenon it is, and with a sample size of 200 enhancers tested, the answer was: pretty common!
Another thing we did not find—and believe me, we looked—was some kind of “silencer state” defined by a combination of chromatin marks that would predict silencer activity. When we scored all the tested elements for occupancy in a range of published histone modification ChIP signal and clustered on these scores, we did see enrichment of silencers in clusters with higher levels of the canonical repressive marks. But membership in these clusters was neither sensitive (lots of silencers fell outside them) nor specific (lots of elements clustered with silencers that showed no evidence of silencer activity, even when tested individually). Groucho enrichment at silencers is modest and not statistically significant. CtBP is depleted at silencers, though again not significantly. Histone deacetylases are modestly depleted, too. We are not ready to start predicting silencers genome-wide. One caveat there is that there are antibodies commercially available (that claim to be ChIP-grade) for about 60 more chromatin marks than we had access to ChIP data for. There could still be a silencer state; we just didn’t find it.
One thing we did see enriched at mesodermal silencers is the transcription factor Snail. By one view this is unsurprising: Snail is the well-studied master repressor of non-mesodermal gene expression in the mesoderm. But it’s also one of the best-studied examples of a short-range repressor. We validated its involvement by mutating Snail binding sites in a few silencers and showing it reduced their activity, at a much longer distance (over 400 bp) than previously associated with short-range repression. We also generated Hi-C data from embryonic cells and saw that silencers that Snail does not bind to are enriched for contacts to promoters. (The Snail-bound silencers are depleted in promoter contacts, but not significantly.) So perhaps we see tentative support for a distinction between short-range and long-range repressor activity, but (as is so often the case in biology) it’s more complicated than we thought.
As biologists, we’ve had a lot of luck with taxonomy over the years, so it makes sense that genomics would start with an attempt to classify the different kinds of sequences in a genome. But we are finding that the distinctions we draw in our minds are a lot sharper than the ones we observe in nature. While we were working on this project, another paper came out5 (and damn near gave me a heart attack) with a title that mentions “Dual functionality of cis-regulatory elements.” This work showed that many enhancers are also Polycomb response elements. Throw in other data blurring the function of insulators with enhancers, and I feel like a picture is emerging where we have to think very carefully about “cis-regulatory elements” with various overlapping subsets of a spectrum of possible regulatory activities.
REFERENCES:
- Jiang, J. et al. Conversion of a dorsal-dependent silencer into an enhancer: evidence for dorsal corepressors. EMBO J. 12, 3201-3209 (1993).
- Gisselbrecht, S. et al. Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos. Nat. Methods 10, 774-780 (2013).
- Gray, S. et al. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes & Dev. 8, 1829-1838 (1994).
- Gisselbrecht, S. et al. Transcriptional silencers in Drosophila serve a dual role as transcriptional enhancers in alternate cellular contexts. Mol. Cell 77, 324-337 (2020).
- Erceg, J. et al. Dual functionality of cis-regulatory elements as developmental enhancers and Polycomb response elements. Genes & Dev. 31, 1-13 (2017).


 (4 votes)
(4 votes)