The people behind the papers: James Nichols
Posted by the Node Interviews, on 29 November 2016
Today’s paper comes from the latest issue of Development, and reveals a link between phenotypic variability, cell fate switching and epigenetic silencing in zebrafish. Lead author James T. Nichols, who carried out the work in Charles Kimmel’s lab in Euegene, Oregon and is now an Assistant Professor in UC Denver’s School of Dental Medicine, gave us the story behind the paper.

Can you tell me your scientific biography leading up to this work? When did you first get interested in craniofacial development?
I first got interested in the skeleton when I was a technician at the biotechnology company Amgen, working on an osteoporosis therapy. After deciding to pursue my own scientific questions I enrolled in graduate school at the University of California, Los Angeles studying Notch signalling in cell culture with Geraldine Weinmaster, that’s where I fell in love with imaging cells. After my Ph.D., I wanted to move from cells in a dish to an in vivo genetic system. Chuck Kimmel’s laboratory at the University of Oregon was the ideal fit for me because studying zebrafish craniofacial development perfectly satisfied my interest in the skeleton, my love of imaging live cells, and powerful vertebrate genetics. Plus, Chuck is the very best mentor anyone could ever ask for, his lab is a really great place to be.
The team behind the paper includes labs from across the States: how did these labs come together for the work?
I love the collaborative nature of modern science. I think that, within the zebrafish community especially, people are eager to share their unpublished findings and reagents to help each other out and that’s what happened in this case. This open communal approach to science is not only more efficient, but also more fun since you get to work with friends from all over the place.
“I love the collaborative nature of modern science… within the zebrafish community, people are eager to share their unpublished findings and reagents to help each other out and that’s what happened in this case.”
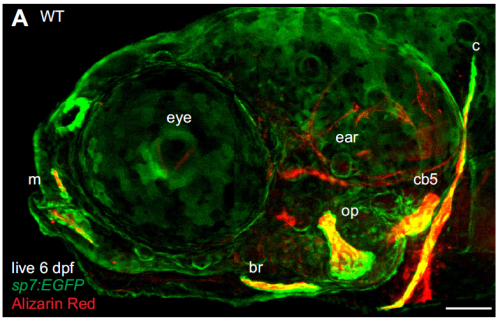
Many researchers, confronted with a highly variable phenotype, would run for the hills or move on to another mutation. What made you guys stick it out?
While this mutant phenotype is notably variable, most mutant phenotypes are more variable than that of the wild type and no one knows why. It is tempting to run for the hills, or just report a ‘representative phenotype’, but I think we are glossing over a lot of interesting biology when we do that. Because zebrafish breed frequently and give large numbers of offspring, and the allele we focused on was behaving just plain weird, we felt like we had a good system to address some important questions about variability. Also, it was very exciting to feel like we were making progress on a real mystery that has intrigued biologists for a long time, I think that kept it fun.
“It is tempting to run for the hills, or just report a ‘representative phenotype’, but I think we are glossing over a lot of interesting biology when we do that.”
Can you sum up the key results of the paper in a paragraph?
We started with an extremely variable gain of bone phenotype that some really smart people in Chuck’s laboratory had made good progress on in the past. However, this mutant had always been problematic because the extra bone phenotype would occasionally disappear. For example, when we moved the mutation onto a transgenic background to label bone cells, the mutant animals would no longer develop extra bone! In this work, we discovered that the ectopic bone was developing because of a transfating event, where bone forms instead of a previously undescribed ligament. We then showed that we could shift the penetrance of transfating up or down through selective breeding, suggesting variability is heritable. When we compared mutants from our selected high- and low-penetrance strains, we found that the high-penetrance strain had more mutant transcripts than the low-penetrance strain. These data suggested that a high level of the mutant transcript was making the phenotype more severe, which we experimentally confirmed by manipulating the transcript. These findings were a real surprise because it suggested that the allele was acting like an antimorph, or a dominant negative, yet it’s completely recessive to the wild-type allele. So it’s an unusual type of allele, a recessive antimorph. We then found that there was a transposon right at the start of the mutant gene, and this transposon was epigenetically different in the two strains. These data suggested that heritable variability in epigenetic silencing might underlie heritable phenotypic variation. Perhaps not just in this context, but in many others as well.
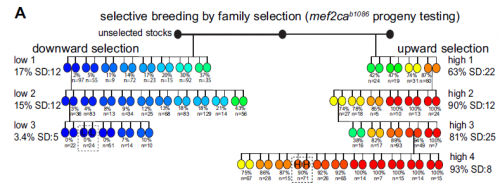
And your selection experiments were based on those done in the early to mid twentieth century? Did you feel like you were addressing the same issues these researchers faced decades ago?
I do, and those studies certainly shaped how we interpreted our results. C.H. Waddington’s work has enjoyed a tremendous resurgence in interest lately, and his genetic assimilation studies really influenced our thinking. Waddington unveiled cryptic variation in his lines that he could selectively breed by heat shocking his flies. We revealed cryptic variation that we could selectively breed by mutagenizing a transcription factor in our fish. Also, the important experiments from Sewall Wright in which he hybridized his inbred guinea pig strains that had an extra toe or not, just like we hybridized our inbred zebrafish strains that had extra head bones or not. Wright interpreted his results as evidence for threshold characters, where if you have enough of some unknown ‘stuff’ you develop a particular phenotype. This led us to look for levels of ‘stuff’ which turned out to be mutant transcripts. It’s so fun to read the old literature and realize that it applies to what we are seeing in the lab; I love the classics!
“It’s so fun to read the old literature and realize that it applies to what we are seeing in the lab; I love the classics!”
Does your work suggest anything about the evolution of new traits?
I think it does, the idea that all this beautiful cryptic variation is just hiding under the surface, unseen in the wild type but ready to be revealed and selected upon is very attractive to me. When conditions are stable, it’s beneficial to have reproducible development. But if conditions change, it’s beneficial to have abundant variation for natural selection to act upon. It’s like having the best of both worlds, consistent development when things are good, but also the ability to provide raw material for evolution when times get tough.
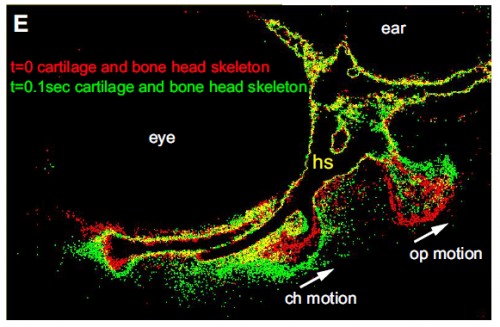
When doing the research, was there a particularly exciting result or eureka moment that has stayed with you?
Absolutely, when we were able to visualize the new ligament for the first time. We suspected it was there, but had no evidence either from us or anywhere in the literature of its existence. Convincing ourselves that it was really there was extremely exciting.
And what about the flipside, any moments of frustration or despair?
We struggled for some time with the concept of a recessive antimorph. How can an allele behave as a ‘dominant negative’ when it’s recessive to the wild type? When we found evidence in the literature from Arabidopsis that this sort of unusual allele exists in other systems it was a big relief.
You’ve just recently started as an Assistant Professor in Denver. How has the transition from Eugene been? What are your plans for the next few years?
I’m living the dream! It took me many years to get to this point and setting up my own laboratory has been everything I’d hoped it would be. There are fantastic colleagues here at the University of Colorado Denver, including some world-class zebrafish laboratories, that have welcomed me and are helping me to get started. Importantly, I have an amazingly supportive wife that enables my science habit. I’m surrounded by wonderful people and that has eased the transition.
“I’m living the dream! I’m surrounded by wonderful people and that has eased the transition.”
In the next few years I plan to follow up on the link between epigenetic variability and mutant phenotype variability. I’ve got some nice preliminary data that strengthens this, plus a whole bunch of results I don’t understand just yet. I’ve got plenty to keep me busy.

What do you like to do when you’re not playing with fish?
When I’m not playing with fish in the laboratory, I’m playing with fish outside the laboratory; I’m an aquarium and pond hobbyist and an avid angler. There are some terrific fly fishing opportunities nearby, and I’m enjoying learning my new home waters.
, , , , , , , , ,
Browse the People Behind the Papers archive here.


 (5 votes)
(5 votes)