The BSDB Beddington Medal Winner 2017: Erik Clark
Posted by BSDB, on 11 April 2017

The BSDB congratulates the 2017 Beddington Medal winner Erik Clark. Erik did his BA in Biological Sciences at the University of Oxford (1st in his year group) and his MSc in Bioinformatics & Theoretical Systems Biology at Imperial College London where he worked on the project entitled “Evolution of Mutation Rate in Fluctuating Environments”. He then moved on to do his PhD within the BBSRC Genes to Organisms Program supervised by Michael Akam at the Department of Zoology, University of Cambridge, where he worked on his project entitled “The Drosophila Pair-Rule System” and where he continues to work now. Erik won an impressive number of prizes, fellowships and grant awards, including the Gibbs Prize in Animal Biology (Univ. Oxford, 2011), an Isaac Newton Trust Research Grant (2016-17), a Junior Research Fellowship (Trinity College, Cambridge) and he is co-investigator on a BBSRC research grant (2017-20).
 His Beddington Medal talk described the outcome of his most successful PhD project. About the background Erik explained: “The Drosophila segmentation cascade is a paradigmatic example of a developmental gene regulatory network, used to gain insight into transcriptional regulation in all animals… Using spatial information from graded domains of “gap” gene expression, seven “pair-rule” genes are expressed in periodic patterns of seven stripes each .. [which] then work in combination to specify precisely-phased 14 stripe patterns of “segment polarity” genes. These output patterns form the template for the segmental organisation of the insect body…[Although] the pair-rule genes have been studied in Drosophila for over 30 years,… there is still no good systems-level understanding of their regulatory interactions“.
His Beddington Medal talk described the outcome of his most successful PhD project. About the background Erik explained: “The Drosophila segmentation cascade is a paradigmatic example of a developmental gene regulatory network, used to gain insight into transcriptional regulation in all animals… Using spatial information from graded domains of “gap” gene expression, seven “pair-rule” genes are expressed in periodic patterns of seven stripes each .. [which] then work in combination to specify precisely-phased 14 stripe patterns of “segment polarity” genes. These output patterns form the template for the segmental organisation of the insect body…[Although] the pair-rule genes have been studied in Drosophila for over 30 years,… there is still no good systems-level understanding of their regulatory interactions“.
In his thesis, Erik used a combination of modelling and experiment to reverse-engineer the structure of the Drosophila pair-rule network and understand how it generates expression dynamics that lead to the patterning of segmental boundaries. He set out to collect a complete time-resolved dataset of relative expression for all pairwise combinations of the 7 pair-rule genes in wild type embryos (see Figure), and a partial dataset for a number of mutant genotypes. Using these data he defined how the regulatory interactions of pair-rule genes change at the mid-cellularisation stage, and identified odd-paired as a temporally regulated factor responsible for these network changes. As an outcome of his work, Erik proposes that that spatial resolution emerges from temporal dynamics, rather than static positional information.
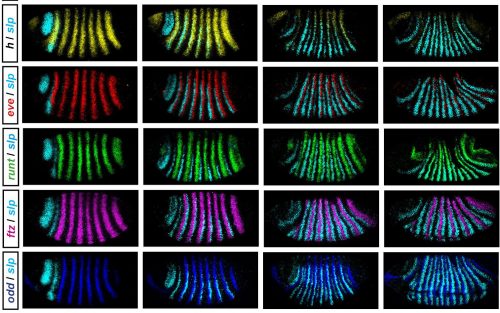
A further part of his thesis proposes that the standard model for how parasegment boundaries are specified, by the interpretation of local gradients of Even-skipped protein, may not be correct. He suggests instead that the shifting of even-skipped stripes across the field of cells in the blastoderm, driven by dynamic gap gene expression, coupled with the temporal control of network interactions, may generate the key offsets in downstream gene expression, and this is an entirely novel idea.
To illustrate Erik’s path to success, Michael Akam writes: “Unlike any other student I have had, Erik spent pretty much the whole of his first year reading. He worked through the entire literature on Drosophila segmentation (spanning 30 years and hundreds of papers), assessing the claims made on the basis of the data presented, and with the advantage of hindsight that the original authors lacked. I suspect he has a more detailed and critical knowledge of this literature than any other researcher.” Michael concludes his support letter with the words: “Erik’s work is strikingly original, and represents a major innovation in thinking about Drosophila segmentation.”
Erik’s publications so far:
- Clark, E. (2017) ‘Dynamic patterning by the Drosophila pair-rule network reconciles long-germ and short-germ segmentation’. bioRxiv: 1101/099671
- Clark E & Akam M (2016). Odd-paired controls frequency doubling in Drosophila segmentation by altering the pair-rule gene regulatory network. eLife: 7554/eLife.18215
- Clark E. & Akam M. Drosophila pair-rule gene double FISH Data (data from Clark & Akam 2016). org:10.5061/dryad.cg35k 2016


 (2 votes)
(2 votes)