BSDB Gurdon/The Company of Biologists Summer Studentship Report #27 – Marketa Novotna
Posted by BSDB, on 19 December 2018
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding. You can read accounts from previous years here.
Our sixth report from the 2018 group of student awardees comes from Marketa Novotna (student at Dundee) who undertook her research with Pauline Schaap (also at Dundee).
Like many other lucky students, I had the chance to participate in real cutting-edge research this summer thanks to the Gurdon Studentship award. Until then, I had spent my time learning the essential theory and mastering various lab techniques. What I was missing, however, was doing actual research that leads to brand new findings, rather than predictable results I’d get in a practical, which had been tried many times before. To me, the summer project represents a transition from only learning a theory and lab techniques to joining a team of scientists in a real-life lab and producing new data, that can advance the field. That is really important to me because contributing to the general pool of knowledge has always been my greatest motivation to study science.
I was hosted by the lab of Prof Pauline Schaap in the University of Dundee. The lab concentrates on several species of slime moulds that are members of the Dictyostelia clade, in particular the model organism Dictyostelium discoideum. These social amoebas are unicellular under normal conditions but environmental stress – especially lack of nutrients or draught – can trigger formation of multicellular fruiting bodies that consist of many hundreds differentiated cells that are derived from the individual amoebas. Some individuals within the structure encapsulate and survive the harsh conditions in form of spores that germinate when environmental conditions improve. The formation of fruiting body is a complex process, which involves intricate cell signalling that ensures a coordinated movement and differentiation of cells.
The long-term mission of the Schaap lab is to understand how this and similar processes evolved from ancestral pathway controlling encystation in more primitive, solitary amoebas and thus partially uncover how multicellularity evolved.
Fruiting body formation in Dictyostelium discoideum
D.discoideum is a species of social amoeba that is unicellular under normal conditions. However, low concentration of food source in a combination with a high density of amoebas in the surrounding environment leads to an exit from the unicellular life cycle. The signals they produce activate PKA (cAMP-dependent kinase), which results in cAMP production. As cAMP diffuses to the environment it acts as a chemoattractant. Individual cells not only respond to this attractant, migrating closer to the source, but they also produce more cAMP, causing pulses of this chemical, which drive more amoebas towards the source. This results in aggregation of individuals that form a mound, which elongates and eventually topples over to create a migrating slug-like structure. Populations of cells start to differentiate into different cell types like pre-spore and pre-stalk cells. (1)
The slug follows environmental cues such as light or warmth to move towards the soil surface. Once it reaches the destination, the cells differentiate further into terminal cell types as the fruiting body develops. Some cells differentiate to form the stalk that serves as a scaffold to hold a mass of differentiated spore cells. A basal disc structure is formed at the base to support the stalk and cells also form upper and lower cups to support the spore head attachment to the stalk. (1)
My project
I worked under a day-to-day supervision of an amazing, patient PhD student Gillian. She’s been studying potential marker genes for distinct parts of the fruiting body (stalk, basal disc, lower cup, upper cup and spores) and the signalling pathways linked to formation of these structures.
Previous work done in the lab identified a number of genes that could play an important role in formation of one of the fruiting body structures due to their enrichment in a specific cell type. Out of these, I studied two genes that looked most promising and went on to establish whether they are expressed in the same parts of fruiting body as hypothesised. For simplicity, I will call them gene A and B.
I used PCR to multiply the promoter sequence of each studied gene then inserted it into a plasmid, which I then used for transformation of E. coli. As the bacteria proliferated, I was able to obtain enough DNA to sequence it and confirm I had the correct sequence. The confirmed promoter sequence could then be inserted into a plasmid I used for transforming D.discoideum. For this purpose, I used a plasmid containing the LacZ reporter gene directly after the promoter-insertion site. This LacZ gene encodes the enzyme β-galactosidase; therefore, since the expression of LacZ was controlled by promoter of the studied gene, the B-gal production mirrored expression of the studied gene.
Next, the gene expression could be visualised using the β-galactosidase substrate, X-gal. After addition of X-gal, a blue precipitate forms at the areas of the fruiting body where the promoter was activated. Hence, allowing us to locate where our gene of interest is expressed and determine whether they are cell-type specific. (2)
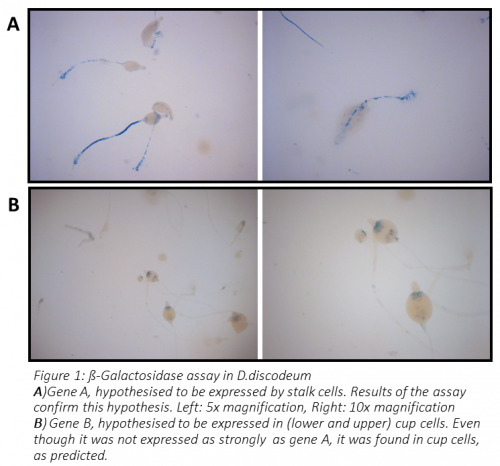
The genes that I worked on – A and B – were hypothesised to be expressed in the stalk and cup, respectively. After I’d spent a great deal of time on optimisation of PCR conditions and several attempts to transform D.discoideum, I acquired transformed amoebas on which I could perform the X-gal staining.
You can see the result of this experiment in Figure 1. In the case of gene A, the stalk was clearly stained, while fruiting bodies of amoebas transformed with gene B promoter showed staining of cup cells (both upper and lower cup). Therefore, the hypothesis was confirmed for both genes.
If I had more time, it would be interesting to find out if these genes are essential for formation of the respective structures by knockout experiments. Furthermore, it could be tested what signalling molecules trigger expression of these genes to further investigate their role in fruiting body formation.
I would like to thank the lab of Prof Pauline Schaap for hosting me and offering a great amount support within a friendly environment. I am also very grateful to my day-to-day supervisor Gillian, who has taught me so much during my placement.
Finally I would like to thank and appreciate the British Society for Developmental Biology for making this experience possible by selecting me for the Gurdon Studentship award. The summer project made me realise that I would really like to pursue a PhD and I would strongly recommend this scheme for any student who is considering a career in science.
References
- Schaap, P. (2011). Evolutionary crossroads in Developmental Biology: Dictyostelium discoideum. Development 138, 387-396.
- Dingermann T, Reindl N, Werner H, Hildebrandt M, Nellen W, Harwood A, Williams J, Nerke K. Optimization and in situ detection of Escherichia coli beta-galactosidase gene expression in Dictyostelium discoideum. Gene. 1989 Dec 28;85(2):353-62.



 (No Ratings Yet)
(No Ratings Yet)