BBSRC DTP: The evolution of nerves: understanding the roots of neurodegeneration
Posted by Andreas Prokop, on 10 November 2022
Location: Manchester
Closing Date: 9 December 2022
How to apply:
- info link: https://www.bmh.manchester.ac.uk/study/research/bbsrc-dtp/projects
- Application deadline: 09 Dec 2022
- Interviews: week commencing 13 Feb 2023
Project description
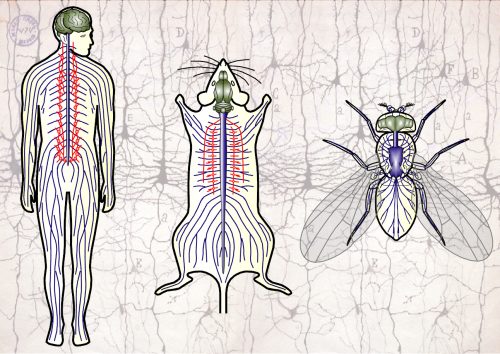 Axons are slender extensions of neurons which can be meter-long and form the biological cables that run through our nerves to hardwire the nervous system. In humans, axons must survive for up to a century; we lose ~40% of axons towards high age and many more in neurodegeneration, but the causes are poorly understood.
Axons are slender extensions of neurons which can be meter-long and form the biological cables that run through our nerves to hardwire the nervous system. In humans, axons must survive for up to a century; we lose ~40% of axons towards high age and many more in neurodegeneration, but the causes are poorly understood.
To understand axon maintenance and pathologies, we are focussing on the bundles of microtubules (MTs) that run all along axons – be it in a tiny fly or in a human [1]. These bundles determine axon structure and form the highways for life-sustaining axonal transport. Accordingly, MT bundle decay causes axon degeneration. But the mechanisms that maintain these bundles (and might fail in pathology!) are little understood [2,3].
On this project, you will test the long-standing, but poorly proven hypothesis that axonal bundles are cross-linked by MT-binding proteins analysing candidate proteins. For example, MAP1B/Futsch proteins are suspected MT cross-linkers, known to be enriched in axons across the animal kingdom with an intriguing evolutionary profile with some parts highly conserved, others extremely variable [1]. You will explore the rules behind this profile and its relevance for axon bundles.
On this project, you will use inter-disciplinary experimental approaches that equip you with a wide range of skills relevant for the biomedical sciences and evolution biology. (a) To determine the precise sub-cellular localisation of MAP1B/Futsch you will use CRISPR/Cas9-mediated protein tagging and apply expansion and/or electron microscopy; molecular mechanisms will be determined via biochemical and in vitro assays. (b) To study MAP1B/Futsch family evolution you will use computational bioinformatics retrieving and analysing Futsch sequences from multiple species. (c) To determine the functional consequences of evolutionary variability you will generate hybrid proteins and assess their impacts on axon architecture.
References
Project info
Location and Supervisors
- Location: Faculty of Biology, Medicine and Health, The University of Manchester
- Supervisors:
- Andreas Prokop has studied the Drosophila nervous system for 30 years, has long-standing experience with electron microscopy, and is an expert in science communication (https://poppi62.wordpress.com/publications).
- Viki Allan studies MT-based transport with expertise in biochemical and in vitro assays to dissect functions of MT-associating proteins [4].
- Matt Ronshaugen specialises on evolutionary biology and his lab is equipped to perform the computational analyses and generate CRISR/Cas9 variants [5].
Funding Notes
Funding will cover UK tuition fee and stipend. We are able to offer a limited number of scholarships that will enable full studentships to be awarded to international applicants. These full studentships will only be awarded to exceptional quality candidates, due to the competitive nature of this scheme.
Closing Date: 9 December 2022
Scientific fields: Cell biology, Neural development
Model systems: Drosophila
Duration: Fixed term
Minimum qualifications: high grade, active project experience

