Fully Funded PhD in Cambridge, UK
Posted by Kathy Grube, on 24 December 2025
Location: Cambridge, UK
Closing Date: 15 February 2026
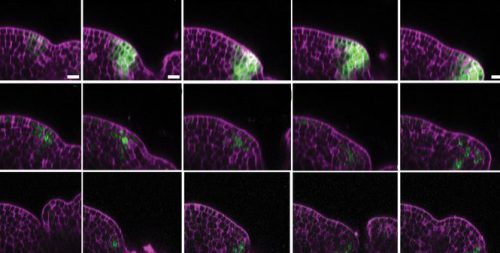
Project Name: Variable and Noisy Gene Expression in the Arabidopsis Shoot Meristem
Institute: Sainsbury Laboratory, University of Cambridge
Supervisors: Professor James Locke (lead supervisor) and Professor Elliot Meyerowitz (co-supervisor)
Enrolment Department: Department of Plant Sciences, School of Biological Sciences, University of Cambridge
Application Deadline: 15 February 2026
Start Date: October 2026
Eligibility: International students are welcome to apply but will need to secure extra financing to cover the remainder of the overseas fees and any immigration-related costs.
Funding: We offer a fully funded studentship, covering fees for Home students and a tax-free stipend at the UKRI rate plus a SLCU 10% uplift for up to four years.
All enquiries should be directed to James.Locke@slcu.cam.ac.uk.
Project Overview
The shoot apical meristem (SAM) is the tiny stem cell niche at the tip of each plant shoot that generates all above-ground organs, stems, leaves and flowers. Its proper function underpins many traits central to crop productivity. Yet even within this tightly organised tissue, gene expression is surprisingly variable from cell to cell. This noise in key developmental regulators may be critical for balancing robustness and flexibility in plant development, but we still do not understand where this variability comes from or how it is controlled.
This project will use state-of-the-art single-cell and spatial transcriptomic approaches to dissect the origins and consequences of gene expression variability in the Arabidopsis SAM. Building on recent single-nucleus RNA-seq work revealing transcriptional heterogeneity in this tissue, you will ask how much of this variability is driven by:
- Cell cycle stage
- Hormonal signalling (e.g. auxin, cytokinin)
- Positional identity within the SAM, captured using spatial transcriptomics and high-resolution imaging
By disentangling these contributions, the project aims to uncover how internal dynamics and spatial cues interact to generate transcriptional noise – and how this noise may be buffered or exploited to maintain robust stem cell function.
Start date: 1 October 2026
Closing Date: 15 February 2026
Scientific fields: Plant development, Quantitative biology and modelling, Signalling, Morphogenesis, Cell biology, Cell fate control and differentiation, Computational and systems biology, Gene regulation, Growth control, Organogenesis
Model systems: Arabidopsis
Duration: Fixed term
Minimum qualifications: Bachelor's degree (e.g. BSc)

