A day in the life of an Oikopleura Lab
Posted by Alfonso Ferrández, on 23 May 2017
The recent bloom of genomic data from all of life’s kingdoms is revealing a novel perspective of gene loss as a pervasive source of genetic variation with a great potential to generate phenotypic diversity and to shape the evolution of gene networks. How do genes become dispensable and subsequently lost? Are patterns of gene loss stochastic or biased? What is the effect of gene loss on the evolution of gene networks? What is the influence of gene loss on the evolution of mechanisms of development (that is, ‘Evo-Devo,) and the diversification of species?
I am Alfonso Ferrández a PhD student working in the group of Prof. Cristian Cañestro and Prof. Ricard Albalat (@EvoDevoGenomeUB) in the Section of Biomedical, Developmental and Evolutionary Genetics in the Department of Genetics (@GeneticsUB), Microbiology and Statistics, and in the Institute of Biodiversity Research (@IRBioUB), of the University of Barcelona, Spain (Fig. 1). We are trying to address some of the previous questions related to gene loss in the field of Evo-Devo using a curious chordate called Oikopleura dioica, which despite having suffered an extreme process of genome compaction along with massive gene losses, still preserves a typical chordate body plan.

Why did we choose O. dioica?

Ecological relevance: O. dioica is a peaceful filter-feeding and free-swimming planktonic organism, about 2-3 mm long. It lives inside a secreted mucous house that it uses as a food trapping device by filtering the water current propelled by its stylish and grooving tail movements (Fig. 2). O. dioica is the only known urochordate species that have separated female and male individuals, which are indistinguishable until maturity. O. dioica has a cosmopolitan world-wide distribution including seas of Europe, Asia and America and it is so abundant in the zooplankton community that plays a key role in marine trophic webs serving as food for fish larvae. Moreover, because of the small size of the pores of their mucous houses, they can also trap the smallest microalgae, thus creating a short circuit that accelerates the transference of organic matter both through the marine trophic web and towards vertical flux of carbon-rich organic material (i.e. marine snow) that sinks to the bottom of the oceans.
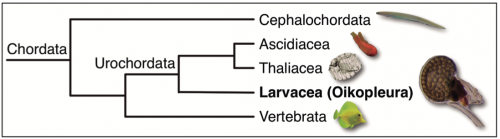
Phylogenetic position within our own phylum: O. dioica belongs to the larvacean class inside the urochordate subphylum, the closest sister group to vertebrates. Urochordates diverged prior to the two rounds of whole genome duplication (2R-WGD) that occurred early in the vertebrate lineage (Albalat and Cañestro, 2016; Cañestro, 2012), and consequently, the mutational robustness of most gene networks appears to be much lower in O. dioica than in vertebrates (Fig. 3). The availability of many deeply sequenced genomes of several other chordates –3 species of cephalochordate, 10 urochordate ascidians, and >100 vertebrates– provides a perfect phylogenetic framework for the identification of gene loss events in O. dioica by comparative genomics with other chordates.
Genomic plasticity: O. dioica has the smallest chordate genome known so far with only 65Mb (even smaller than the 175 Mb of Drosophila or the 100 Mb of C. elegans), which results from an extensive process of compaction that has been accompanied by an extraordinary amount of gene losses. One striking example is the loss of all genes of the non-homologous end joining DNA repair system, plausibly one of the reasons that accounts for the elevated propensity for gene loss of this organism. Among key developmental genes, O. dioica has lost more than 30% of the homeobox gene groups, including all central Hox genes, and key genes involved in retinoic acid (RA) signaling (Albalat and Cañestro, 2016; Cañestro et al., 2006; Denoeud et al., 2010; Edvardsen et al., 2005; Martí-Solans et al., 2016; Seo et al., 2004).
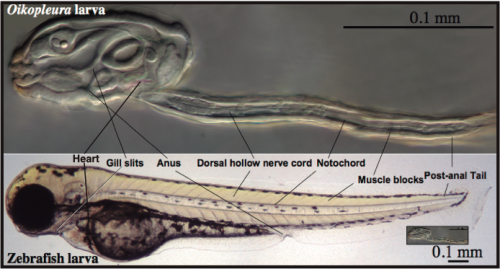
A simple and transparent model for Developmental Biology: The embryonic development of O. dioica is very fast, and in less than 20 hours a transparent juvenile already shows a typical chordate body plan with organs that are unequivocally homologous to those in vertebrates, including a notochord anchoring muscle cells throughout a post-anal tail, a dorsal neural tube, brain, thyroid, pituitary, gill slits, pharynx, esophagus, gut and a heart (Fig. 4). In addition, O. dioica shares with other urochordate species, such as ascidians, a very similar embryonic developmental program, both at the morphological and molecular level, with the important difference that O. dioica does not suffer the drastic metamorphosis that ascidians do, and therefore, in contrast to ascidians, maintain all chordate features throughout its life.
A day in the life of an Oikopleura lab
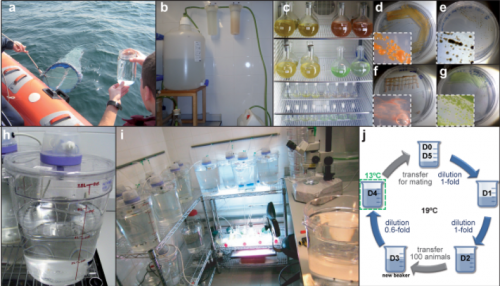
Our laboratory is one of the few laboratories in the world able to culture O. dioica all year round, which means almost unlimited availability of biological material (i.e. mature males and females, eggs, sperm, embryos and larvae) to carry out functional experiments. We maintain them at 19ºC, which results in a generation time of only 5 days (Fig. 5). Our lab has set up a low-cost maintenance regime by reducing as much as possible the amount of water, space and manpower to handle the animals (Martí-Solans et al., 2015). All these characteristics, together with its high fecundity and transparency makes it an attractive model for developmental studies.
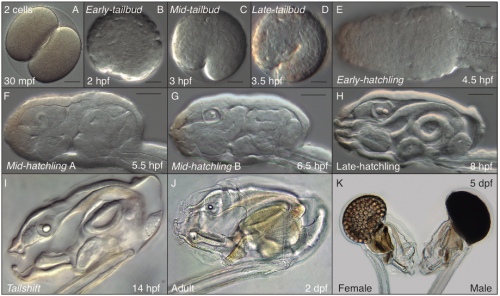
A typical day in the Oikopleura lab starts looking for ripe animals to mate and start a new generation (this happens early in the morning, since we have synchronized the animals to mature at that time of the day). At day 5 of their cycle, males and females are easily distinguishable. Males have a yellowish gonad full of sperm, whereas females have a refringent and translucent gonad full with 100 to 400 eggs (Fig. 6 K). Since O. dioica has external fertilization, to mate them we only have to put together about 20 females and 10 males, and wait for their spontaneous spawn that will give rise to the next generation. Both males and females naturally die after the spawning, which unfortunately does not allow us to keep the parental generations. Next day (day 1), we are normally happy to see hundreds of juveniles beating their tales inside their already inflated houses. Yes! They have a brisk development! The first division occurs just 20 minutes after fertilization; by 4 hours after fertilization (hpf) they break their chorion, and few minutes after the hatch we can already see them graciously twitching their tails in their first attempts to swim. By 5 hpf the tail movements are rhythmic and harmonious, which allow them to swim up and being suspended in the column of water. By 8 hpf, the heart is vigorously beating, and the ciliary rings are working at full speed creating the water to circulate through the pharynx. By 9 hpf, the animals do the tailshift, characterized by the shift of the tail to an acute angle relative to the trunk, flagging the end of embryonic development, and competence to secrete and inflate their first filter-feeding house (Fig. 6).
From day 1 to day 5, we need to feed them every single day (weekends and bank holidays included, aggh!), and to transfer them to fresh seawater to keep them happy, and not too crowded. Their diet consists of a mix of four different species of algae that we also grow in the lab, at different ratios depending on the size and needs of the animals each day of the culture (Fig. 5 c-g).
Functional approaches of gene knockdown or inhibition are amenable. The syncytial gonad of females is easy to inject with RNAi, morpholinos or even, the recent discovered DNAi (yes, dsDNA rather dsRNA of your target gene…cheap and effective), obtaining a massive generation of knockdown embryos (Omotezako et al., 2015). Moreover, permeability and small size of O. dioica embryos allow us to easily treat them with drugs or specific inhibitors of signaling pathways to modify their developmental programs.
Current research lines
To address the fundamental question of how gene loss affects the evolution of the mechanisms of development, as a case study, our research focuses on the analyses of the striking loss in O. dioica of the retinoic acid (RA) signaling pathway, which is conserved and essential for many developmental and physiological roles in all other known chordates. First, we have described a process of gene co-elimination of nearly the entire classic metabolic and signaling pathways. This analysis allowed us also to recognize surviving genes to the dismantling of those pathways, and to recognize processes of neofunctionalization and hidden pleiotropy as the probable causes that preserved the genes of vanishing. Currently, our focus of attention is on O. dioica heart development, since RA plays a fundamental role in all other chordates. Finally, two new lines of applied research are starting to fly in our lab, using O. dioica as an evolutionary knockout model to study the genetic bases of some human cardiomyopathies, as well as a model to better understand the limits of the genetic responses of chordate development towards environmental threats from anthropogenic origin such as heavy metal from industrial wastes or global warming (but these are two long new stories that would need another thread in the Node).
References
Albalat, R., Cañestro, C., 2016. Evolution by gene loss. Nat. Rev. Genet. doi:10.1038/nrg.2016.39
Bouquet, J.M., Spriet, E., Troedsson, C., Otter, H., Chourrout, D., Thompson, E.M., 2009. Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica. J. Plankton Res. 31, 359–370. doi:10.1093/plankt/fbn132
Cañestro, C., 2012. Two Rounds of Whole-Genome Duplication: Evidence and Impact on the Evolution of Vertebrate Innovations, in: Polyploidy and Genome Evolution. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 309–339. doi:10.1007/978-3-642-31442-1_16
Cañestro, C., Postlethwait, J.H., Gonzàlez-Duarte, R., Albalat, R., 2006. Is retinoic acid genetic machinery a chordate innovation? Evol. Dev. 8, 394–406. doi:10.1111/j.1525-142X.2006.00113.x
Denoeud, F., Henriet, S., Mungpakdee, S., Aury, J.-M., Da Silva, C., Brinkmann, H., Mikhaleva, J., Olsen, L.C., Jubin, C., Cañestro, C., Bouquet, J.-M., Danks, G., Poulain, J., Campsteijn, C., Adamski, M., Cross, I., Yadetie, F., Muffato, M., Louis, A., Butcher, S., Tsagkogeorga, G., Konrad, A., Singh, S., Jensen, M.F., Huynh Cong, E., Eikeseth-Otteraa, H., Noel, B., Anthouard, V., Porcel, B.M., Kachouri-Lafond, R., Nishino, A., Ugolini, M., Chourrout, P., Nishida, H., Aasland, R., Huzurbazar, S., Westhof, E., Delsuc, F., Lehrach, H., Reinhardt, R., Weissenbach, J., Roy, S.W., Artiguenave, F., Postlethwait, J.H., Manak, J.R., Thompson, E.M., Jaillon, O., Du Pasquier, L., Boudinot, P., Liberles, D.A., Volff, J.-N., Philippe, H., Lenhard, B., Roest Crollius, H., Wincker, P., Chourrout, D., Delsuc, F., Brinkmann, H., Chourrout, D., Philippe, H., Bouquet, J.M., Eickbush, T.H., Furano, A. V., Volff, J.N., Lehrach, H., Reinhardt, R., Chourrout, D., Woolfe, A., Nguyen, H.D., Yoshihama, M., Kenmochi, N., Marz, M., Kirsten, T., Stadler, P.F., Zorio, D.A.R., Blumenthal, T., Mourier, T., Jeffares, D.C., Roy, S.W., Gilbert, W., Roy, S.W., Irimia, M., Cousineau, B., Li, W., Tucker, A.E., Sung, W., Thomas, W.K., Lynch, M., Edvardsen, R.B., Satou, Y., Satoh, N., Hughes, T., Liberles, D.A., Thompson, E.M., Kallesøe, T., Spada, F., Katju, V., Lynch, M., Putnam, N.H., Putnam, N.H., Srivastava, M., Srivastava, M., Lynch, M., Conery, J.S., Lynch, M., Lynch, M., 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330, 1381–5. doi:10.1126/science.1194167
Edvardsen, R.B., Seo, H.C., Jensen, M.F., Mialon, A., Mikhaleva, J., Bjordal, M., Cartry, J., Reinhardt, R., Weissenbach, J., Wincker, P., Chourrout, D., 2005. Remodelling of the homeobox gene complement in the tunicate Oikopleura dioica. Curr. Biol. doi:10.1016/j.cub.2004.12.010
Martí-Solans, J., Belyaeva, O. V., Torres-Aguila, N.P., Kedishvili, N.Y., Albalat, R., Cañestro, C., 2016. Co-elimination and survival in gene network evolution: dismantling the RA-signaling in a chordate. Mol. Biol. Evol. 33, msw118. doi:10.1093/molbev/msw118
Martí-Solans, J., Ferrández-Roldán, A., Godoy-Marín, H., Badia-Ramentol, J., Torres-Aguila, N.P., Rodríguez-Marí, A., Bouquet, J.M., Chourrout, D., Thompson, E.M., Albalat, R., Cañestro, C., 2015. Oikopleura dioica culturing made easy: A Low-Cost facility for an emerging animal model in EvoDevo. Genesis 53, 183–193. doi:10.1002/dvg.22800
Omotezako, T., Onuma, T.A., Nishida, H., 2015. DNA interference: DNA-induced gene silencing in the appendicularian Oikopleura dioica. Proc. R. Soc. B 282, 20150435. doi:10.1098/rspb.2015.0435
Seo, H.C., Edvardsen, R.B., Maeland, A.D., Bjordal, M., Jensen, M.F., Hansen, A., Flaat, M., Weissenbach, J., Lehrach, H., Wincker, P., Reinhardt, R., Chourrout, D., 2004. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431, 67–71. doi:10.1038/nature02709


 (2 votes)
(2 votes)
Hola desde México ! Para una persona no familiarizada con el tema, como establecen que este organismo tuvo perdida de genes ? Comparando el tamaño de su genoma con el tamaño del genoma de otros organismos relacionados en la escala filogenética ? Leí su trabajo de “Evolution by gene loss” y solo entendí un 60 % aprox. lo que no me impide reconocer su trabajo y dedicación. Adelante y muchas felicidades.