MorphoGraphX: A platform for quantifying morphogenesis in 4D
Posted by Anne-Lise Routier-Kierzkowska, on 15 June 2015
Quantifying shape, growth and gene expression at the cellular level are key to understanding morphogenesis, i.e. how organs are shaped. Many image processing tools have been developed towards this goal that operate on either 2D or 3D images. 2D tools are fast, easy to use, and typically involve datasets of modest size. However organs and tissues are often highly curved and a single 2D image can be not sufficient to quantify their shape. To address this problem, 3D tools have been developed. Yet these require very large and high quality images, with deep penetration that is often not possible with live imaging. We have developed the image analysis software MorphoGraphX1, that gets around these problems by working with curved 2D surface images. We like to call it image processing in “2.5D”.
MorphoGraphX was initially developed to answer specific questions in plant biology. A particularly challenging one was: Is there a difference in mechanical properties between the shoot apex stem cell niche and the surrounding cells2? To answer this question we used osmotic treatments to lower the cell internal pressure, with the idea to precisely track the shape change of the cells. Softer cells would be expected to deform more. However this approach presented a major technical problem. We needed to very precisely track small changes (~8-10% in area) in cell size on the surface of the shoot apex from 3D confocal image stacks. Due to the curvature of the apex, 2D sections would not work, and the image quality was not sufficient to do a full 3D segmentation, in particular because osmolytes contained in the medium scattered light.
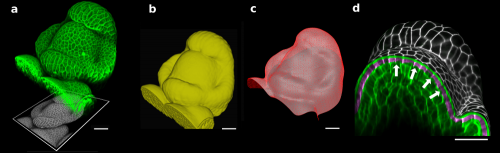
Figure 1. The 3D confocal stack of an arabidopsis flower (a, in green) shows how curved many biological shapes can be. In this case, a flat projection of the 3D data (a, in grey) leads to too much distortion and cannot be used to study cell shape or quantify signal intensity. Instead, we first extract the global organ shape (b) from the 3D data and create a curved triangular mesh (c) that represents the surface. A narrow band of the 3D image data (d, purple) can then be projected onto the surface, creating a curved (2.5D) image of the cell outlines (here is grey scale). Scale bars: 20 μm. Adapted from (1).
To solve this problem, Richard Smith’s team had the idea of first extracting the global shape of the apex and then projecting the cell outlines onto this curved surface, creating a curved 2.5D image of the surface. This way, cells on the flanks of the organ would not be distorted by the projection. The process proved to be robust enough to extract relatively small changes in cell shape under difficult imaging conditions2.
Quickly it became apparent that the idea could be applied to other biological problems, both in plant and animal research. MorphoGraphX grew into a collaborative project, initially involving several groups in the Swiss SystemsX consortium, the “Plant Growth”, “Wing-X” and “SyBIT” projects. Today MorphoGraphX integrates code and algorithms from many different sources, and is used as a fully 3D image processing platform that can be extended in much the same way as ImageJ or Fiji. One key difference is that processes in MorphoGraphX (plugins) are written in C++, and much of the 3D image processing exploits the latest generation of graphics cards, which can have 1000s of computing cores. Combined, the platform gives the scientist the ability to integrate a variety of tools to create work-flows, test new ideas quickly, and visualize the results easily.
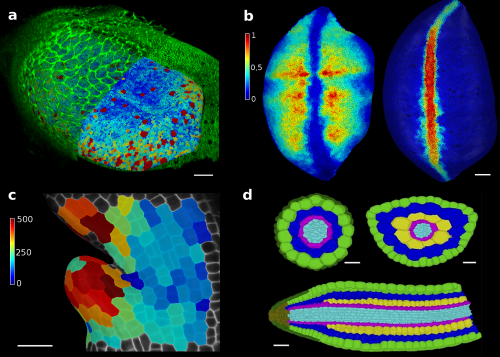
In our paper recently published in eLife1, we show how the tools developed in MorphoGraphX can be applied to solve various problems in development (Figure 2). Among others applications, MorphoGraphX has been used to investigate mechanical properties in the shoot apex2, quantify cell shape in Drosophila wing disk3, dissect how leaf shape emerges from gene expression and growth at the single cell level4, and automatically classify cells according to their tissue type in Arabidopsis embryo5. MorphoGraphX output can be also be used as a templates for numerical simulations in both 2D and 3D, to model hormone transport in roots6, cell division rules in developing embryos7 or mechanical stresses during germination8.
Figure 2. MorphoGraphX tools have proved to be valuable for image analysis in both plant and animal research. The surface of a Drosophila wing disk extracted after digitally removing the peripodial membrane from a 3D confocal stack (a), enabling researchers to analyze the cell shape and size (a) or transcription levels of genes (b), such as vestigial (left) and wingless (right). The software can also be used to track growth of individual cells, for example to analyse the effects of a growth-repressing gene in Arabidopsis leaves. Colorscale represents expansion over 24h in % (c). Cells of an Arabidopsis embryo are colored according to their tissue type. The classification was performed automatically, based cell shape and position extracted from 3D segmentation (d). Scale bars: 20 μm (a and b), 30 μm (c) and 20 μm (d). (a,b) adapted from (1), (c) from (4), (d) from (5).
We use MorphoGraphX in our own lab on a daily basis and collaborate with biologist from various labs around the world. With much of the code developed by people who actually use the software, the emphasize has been on the ease of use, speed, reliability, and the ability to combine algorithms from many sources. Our aim is to share our platform with anyone that finds it useful, and to integrate contributions from the community that are developed in the context of a a wide range of research goals.
For more informations, please visit: www.MorphoGraphX.org and the Smith lab website.
1. Barbier de Reuille, P., Routier-Kierzkowska, A., Kierzkowski, D., Bassel, G., Schüpbach, T., Tauriello, G., Bajpai, N., Strauss, S., Weber, A., Kiss, A., Burian, A., Hofhuis, H., Sapala, A., Lipowczan, M., Heimlicher, M., Robinson, S., Bayer, E., Basler, K., Koumoutsakos, P., Roeder, A., Aegerter-Wilmsen, T., Nakayama, N., Tsiantis, M., Hay, A., Kwiatkowska, D., Xenarios, I., Kuhlemeier, C., & Smith, R. (2015). MorphoGraphX: A platform for quantifying morphogenesis in 4D eLife, 4 DOI: 10.7554/eLife.05864
2. Kierzkowski, D., Nakayama, N., Routier-Kierzkowska, A., Weber, A., Bayer, E., Schorderet, M., Reinhardt, D., Kuhlemeier, C., & Smith, R. (2012). Elastic Domains Regulate Growth and Organogenesis in the Plant Shoot Apical Meristem Science, 335 (6072), 1096-1099 DOI: 10.1126/science.1213100
3. Aegerter-Wilmsen, T., Heimlicher, M., Smith, A., de Reuille, P., Smith, R., Aegerter, C., & Basler, K. (2012). Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size Development, 139 (17), 3221-3231 DOI: 10.1242/dev.082800
4. Vlad, D., Kierzkowski, D., Rast, M., Vuolo, F., Dello Ioio, R., Galinha, C., Gan, X., Hajheidari, M., Hay, A., Smith, R., Huijser, P., Bailey, C., & Tsiantis, M. (2014). Leaf Shape Evolution Through Duplication, Regulatory Diversification, and Loss of a Homeobox Gene Science, 343 (6172), 780-783 DOI: 10.1126/science.1248384
5. Montenegro-Johnson, T., Stamm, P., Strauss, S., Topham, A., Tsagris, M., Wood, A., Smith, R., & Bassel, G. (2015). Digital Single-Cell Analysis of Plant Organ Development Using 3DCellAtlas The Plant Cell, 27 (4), 1018-1033 DOI: 10.1105/tpc.15.00175
6. Santuari, L., Scacchi, E., Rodriguez-Villalon, A., Salinas, P., Dohmann, E., Brunoud, G., Vernoux, T., Smith, R., & Hardtke, C. (2011). Positional Information by Differential Endocytosis Splits Auxin Response to Drive Arabidopsis Root Meristem Growth Current Biology, 21 (22), 1918-1923 DOI: 10.1016/j.cub.2011.10.002
7. Yoshida, S., Barbier de Reuille, P., Lane, B., Bassel, G., Prusinkiewicz, P., Smith, R., & Weijers, D. (2014). Genetic Control of Plant Development by Overriding a Geometric Division Rule Developmental Cell, 29 (1), 75-87 DOI: 10.1016/j.devcel.2014.02.002
8. Bassel, G., Stamm, P., Mosca, G., Barbier de Reuille, P., Gibbs, D., Winter, R., Janka, A., Holdsworth, M., & Smith, R. (2014). Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo Proceedings of the National Academy of Sciences, 111 (23), 8685-8690 DOI: 10.1073/pnas.1404616111


 (4 votes)
(4 votes)