Why do we need to understand the structure of the Enteric Nervous System?
Posted by Reena Lasrado, on 16 June 2017
Insight into the organizational structure of a growing tissue is imperative for understanding its development and function. Structure can reveal the systematic steps undertaken towards making specific positional and cell fate choices/decisions. A well-defined structure helps to dissect the complexity underlying the networks that form as the tissue develops. It helps to elucidate the foundation on which the components can work together within the confines of the system. It helps one to read the lines of communication that are built into the system for its proper functioning. When an organized structure regulates the flow of information, then changes within that information are easier to monitor and adapt.
Historical views
The Enteric Nervous System (ENS) is one of the largest subdivisions of the peripheral nervous system. It earns its’ name as the “second brain” due to the millions of neurons it contains and the complex neural networks it displays1. Although Bayliss and Starling’s description of ‘the law of the intestine’ hallmarked the functional discovery of the ENS already more than a century ago,2 their discovery also unveiled some daunting scientific challenges, many of which are explained by inadequate insight into ENS structure and how this contributes to its function.
Simply put – It is a very challenging system for various different reasons. The ENS is challenged every day with the variety of food/drink we intake, which means the environment is under constant change. The ENS has a mind of its own! It autonomously controls functions of the gut that include peristalsis, secretion of enzymes and absorption of food. The ENS is layered in close apposition to contractile sheets of smooth muscle cell syncytia, thereby complicating several experimental approaches to a large extent. The gut lumen is host to a vast ocean of microbes with which the ENS interacts3 with other systems within this tissue, such as, the immune and epithelial systems to maintain a healthy gut. In addition, it also communicates with the brain to keep the body in good health.
The ENS originates from neural crest-derived progenitors that travel through different spatio-temporal environments as they expand in number, colonize the gut tissue and differentiate to generate a plethora of neuronal and glial cells. Unlike the central nervous system, neurons and glial cells of the ENS do not show any apparent order of arrangement. Cells coalesce in a salt and pepper manner to form ganglia and create an expansive network. However, reproducible patterns of secretory and motor function appear to be almost exclusively controlled by the ENS. Being indispensable for gut physiology, its lack of function is implicated in the pathogenesis of several gastrointestinal disorders, such as Hirschsprung disease and others that are of unknown aetiology.
Important questions
For a system that is tremendously interactive in an extensively expanding tissue (8 metres in humans), knowledge of how its structure develops is very important. This information can be useful in understanding diseases of the gut that may arise during development or later in adulthood and disrupt the network. Therefore, it was imperative for us to make sense of the ‘randomness’ that the ENS displays. We wished to gain an understanding of any fundamental rules that defined its apparent chaotic cellular topology and eventually its contribution to function.
Previous work in many labs including ours, have aimed to understand ENS structure and function by using global approaches and studying it at the population level. However, such approaches have not been very successful in gaining detailed insight into how the lineages of the ENS develop and contribute to its structure and function. As a lab that carries a reputation of developing tools and techniques that help to answer big over-arching questions in the field, we took the plunge. We decided to opt for single-cell approaches so as to address the contribution of an individual progenitor to the spatial development and function of the ENS at both the cellular and molecular level.
The work
Appropriate combination of genetic tools4 were generated in the lab and employed to track the static behaviour of individual ENS progenitors in a multi-colour mosaic manner over the developmental time line. Having optimized the various steps involved in the different experiments our results looked promising enough for me to spend hours and days to localize and image the beautiful clones we obtained in our study. The scanned clones were analysed for their composition, measured left, right and centre to gain an understanding about their spatio-temporal characteristics. We combined in vivo and ex-vivo approaches to capture the cellular properties of these cells. The days spent away from imaging were invested in isolating single cells for the transcriptomic study. This analysis was performed to understand the intrinsic properties of heterogeneous individual progenitors and how ENS lineages are generated. We also used mosaic mutagenesis to disrupt the system during development and understand its effect on ENS composition.
Making sense
With this arduous approach the mysterious topology of the ENS unfolded before our eyes. The close spatial relationship of the labelled families both at the 2D and 3D axes with a single colour meant ‘something’ to the system. This study helped us to unravel a set of rules that define the columnar organization of overlapping clonal lineages. This suggested to us that this structural organization could help the components of the ENS to work and face the challenges of the system together as a family. We also observed that as the system developed, subpopulations emerged in a defined manner. Neurons related to each other showed co-ordinate activity upon stimulation, highlighting their means of communication within the ENS of the small intestine. Molecular analysis of ENS progenitors revealed the manner in which neuronal and glial lineages arise. Further, our mutagenesis study revealed the specific role of an important receptor tyrosine kinase, RET in neuronal commitment. Together, our work suggests that lineage relationships are fundamental for the spatial organization and function of the ENS.
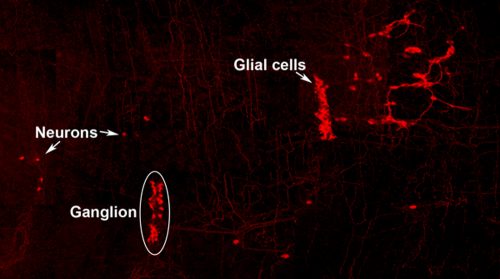
Neuro-glial clone: This image shows the progeny of a neuro-glial progenitor in the myenteric plexus of the adult mouse. Neurons extend processes forming a meshwork of connectivity and glial cells are observed within ganglia.
Future
Perhaps mistakes in the blueprint of the ENS are the cause of gastrointestinal diseases with unknown aetiology. We are yet to uncover the principles that underlie information processing, which will help us to assign the logic of ENS assembly and connectivity. Now that we have a better understanding of how the ENS of the small intestine assembles and underpins its function, we can start to probe this system at different stages of development.
This blog is contributed by Reena Lasrado and Werend Boesmans.
References
- Bayliss WM, and Starling EH. The movements and innervation of the small intestine. The Journal of Physiology 24: 99-143, 1899.
- Gershon MD. The Second Brain. New York, NY: HarperCollinsPublishers, 1998, p. xvi, 314 p.
- Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, Pettersson S, Pachnis V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron. 2015 Jan 21; 85(2):289-95. doi: 10.1016/j.neuron.2014.12.037. Epub 2015 Jan 8.
- Lasrado R, Boesmans W, Kleinjung J, Pin C, Bell D, Bhaw L, McCallum S, Zong H, Luo L, Clevers H, Vanden Berghe P, Pachnis V. Lineage-dependent spatial and functional organization of the mammalian enteric nervous system. Science. 2017 May 19; 356 (6339):722-726. doi: 10.1126/science.aam7511


 (1 votes)
(1 votes)