Plant “Velcro” holds it all together
Posted by dllewellyn, on 27 June 2016
Plant hairs or trichomes mean little to most people until they bite into a furry skinned peach or prick their finger on a rose bush thorn, but in the plant kingdom these versatile epidermal structures perform many essential functions that are attributable to their physical shape, location, density and sometimes chemical composition. Next time you pop on a cotton T-shirt think about where the fibres for that textile come from – it’s made from a plant trichome.
For the last 20 years or more our lab has been concerned with understanding the molecular basis for the differentiation of the long hairs or seed trichomes that develop on the seeds of the cotton plant, Gossypium hirsutum L.. These fibres can become very long, reaching up to 5 cm or more in some cotton species, so could be some of the longest cells in the plant kingdom. They are single cells of the cotton ovule epidermis that elongate and fill with a thick secondary cell wall layer that is almost pure cellulose which imparts the special feel and fabric properties of cotton textiles. Using cDNA microarrays we were able to compare the genome wide expression in cotton ovules just initiating the production of fibres and with the gene expression in ovules from a mutant plant that didn’t make any fibres (Wu et al., 2006). This identified members of two specialised classes of plant-specific transcription factors: MIXTA-like MYB genes and homeodomain-leucine zipper IV (HD-zip) genes that were reduced in expression in the fibreless mutant. These two families of genes appear to have evolved diverse functions in regulating epidermal cell differentiation in plants, including plant trichomes.

Detailed functional analysis of our three candidate genes in transgenic cotton plants using over-expression and gene silencing strategies indicated that the transcription factors MYB25, MYB25-like and HD-1 all played essential roles in cotton fibre development and MYB25 and HD-1 were also key components in the development of other hairs that covered the leaves and stems of the cotton plant (Machado et al., 2009, Walford et al., 2011 and 2012). The sequencing of the cotton genome (Paterson et al., 2012; Zhang et al., 2015) a few years ago (cotton is a polyploid species with two genomes A and D derived from the hybridisation of two ancestral Gossypium diploid species a couple of million years ago) enabled us to look more closely at a whole genome level at all the potential members of these specialised classes of genes. We have focussed mainly on understanding the MIXTA-MYBs. Cotton has ten (including MYB25 and MYB25-like) of these types of genes (twenty if you count both sub-genomes) – seven more than the model plant species Arabidopsis, so there has been an expansion of this gene type in cotton presumably to cope with the large variety of epidermal cell types in cotton, including the seed fibres that are a special feature found in only some plant species!
Over the last few years we have been working our way through the cotton MIXTA-MYB family trying to define their functions using the same approach of characterising their tissue and developmental gene expression patterns and silencing and over-expressing them in transgenic cotton plants. All of them except perhaps MYBML10 (MYB MIXTA-Like gene 10) appeared to be expressed predominantly in cotton ovules during the stages of early fibre development (Bedon et al., 2014). Some of the other genes appear to be part of the regulatory cascade controlling fibre development and are downstream of MYB25-like, but they may also be shared components of development pathways for other types of trichomes. That is another story, but this blog is mainly about MYBML10 that was found to be the master regulator for a very special type of trichome present on the petals of the cotton flower. These trichomes have an interesting evolutionary function and their analysis formed the basis for our recent Letter to Nature Plants (Tan et al., 2016).
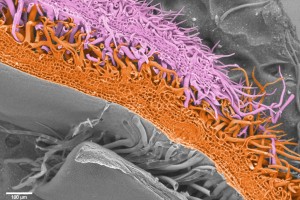
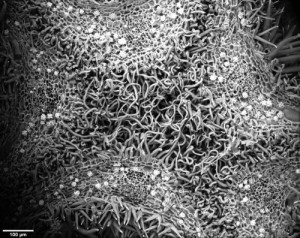
In a phylogeny of the cotton MIXTA-MYB protein sequences, MYBML10 clustered together with MYB25 and MYB25-like, so we expected it to be part of the fibre development pathway, however it was very lowly expressed in whole cotton ovules and in the outer integument layer of the ovule (a 3-4 cell layer that includes the epidermis layer where the fibres initiate and which we can painstakingly dissect away from the rest of the ovule for fine gene expression analysis (Bedon et al., 2013)). When we silenced this gene using RNAi there didn’t appear to be any effect on cotton fibre production, but the sharp eyes of postdoc, Jiafu Tan, noticed that the flower buds of the most silenced lines were abnormal and the seed set on those plants was reduced, mainly because the petals did not cover the anthers and stigma properly when they were very young and parts of these tissues dried out and died. He embarked on a detailed microscopic and molecular analysis to understand why this might be the case and in doing so stumbled on a new type of trichome in cotton that had a pretty important function of holding together the petals of the developing cotton bud to protect the immature reproductive tissues until they were ready to be exposed to the outside environment. Both the outside and inside faces of the cotton petals had trichomes with those on the outside being mostly four-celled stellate or star-shaped trichomes and those on the inside being single celled trichomes. When the petals are folded around each other, as they are in the young flower bud, the outer and inner faces overlap and the two types of trichomes become juxtaposed and entangled just like ‘Velcro’ to lock the petal edges together (false colour picture at top and black and white image below).

Silencing the transcription factor MYBML10 inhibited the development of the trichomes on both faces of the petal and this allowed the petals to slide past one another as they expanded inside the bud causing the bud to twist and expose the young anthers and stigma to the air. As these reproductive tissue were damaged and cotton is a self-pollinating plant, then few seeds were set, so these trichomes were a critical evolutionary development in cotton. By collaborating with a materials science engineer at a neighbouring university’s Engineering Department we were able to take a couple of these petals that were linked together by their trichomes and actuially measure the force needed to pull them apart in a specially designed tension meter – it was a wopping 1 N per petal edge, so these trichomes can withstand amazing forces that must be being generated within the flower bud as the petals expand inside. No wonder the flowers are twisted and abnormal when they don’t have petal trichomes.
Are these petal trichomes something unique to cotton? When we started looking, the answer was a resounding no. Visiting a local nursery (fortunately it was summer), we were able to find a selection of flowering plants to look at. Petal trichomes were by no means found in every flower we looked at, but they were certainly prevalent in the Malvaceae, the plant family that includes cotton and its Hibiscus relatives, but they were also present in a number of other plant families and it wasn’t just on the petals, but also the next outer layer of the flower, the sepals and even the next layer, the bracts.
Petal trichomes have therefore been an important development during plant evolution, particularly in species, like those in the Malvaceae whose large flowers expand rapidly before the reproductive organs are fully adapted to drier environments. Trichomes have also been found in some other species like tomato to hold together separate anthers into something like a salt shaker (anther cone) to allow the pollen to all be released at one time when an insect visits a flower (Glover et al., 2004) so trichomes are one form of Nature’s glue to effectively bind organs together to create the diversity of structures we appreciate as flowers, and while we didn’t find a new gene that we could manipulate to improve cotton fibre production we did unearth some fascinating biology that illustrates the simple beauty of Nature’s engineering.
References
Bedon F, Ziolkowski L, Osabe K, Venebles I, Machado A and Llewellyn D (2013) Separation of integument and nucellar tissues from cotton ovules (Gossypium hirsutum L.) for both high- and low-throughput molecular applications. BioTechniques 54, 44-46.
Bedon F, Ziolkowski L, Walford S-A, Dennis ES and Llewellyn DJ (2014) Members of the MYBMIXTA-like transcription factors may orchestrate the initiation of fibre development in cotton seeds. Frontiers in Plant Science 5, e179 doi:10.3389/fpls.2014.00179
Glover BJ, Bunnewell S and Martin C (2004) Convergent evolution within the genus Solanum: the specialised anther cone develops through alternative pathways. Gene 331, 1-7.
Machado A, Wu Y, Yang Y, Llewellyn D and Dennis ES (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. The Plant Journal 59, 52-62.
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo M-J, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee T-H, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashfari H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong J-K, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MFS, Waghmare VN, Walford S-A, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KFX, Peterson DG, Rokhsar DS, Wang X and Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable fibers. Nature 492, 423–427.
Tan J, Walford S-A, Dennis ES and Llewellyn DJ (2016) Trichomes control flower bud shape by linking young petals together. Nature Plants 2, Article number: 16093. Published online June 20 2016. http://dx.doi.org/10.1038/nplants.2016.93
Walford SA, Wu YR, Llewellyn DJ and Dennis ES (2012) Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant Journal 71, 464-478.
Walford S, Wu Y, Llewellyn DJ and Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. The Plant Journal 5, 785-797.
Wu Y, Machado A, White RG, Llewellyn DJ and Dennis ES (2006). Expression profiling identifies genes expressed during early lint fibre initiation in cotton. Plant and Cell Physiology 47, 107-127.
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski C, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride R, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, and Chen ZJ (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fibre improvement. Nature Biotechnology 33, 531–537 doi:10.1038/nbt.3207


 (6 votes)
(6 votes)