silicoCROSS: a help in genetic crosses
Posted by Mario Metzler, on 12 August 2013
I’m quite a lazy person, and as such I like to find solutions to boring and repetitive tasks.
One of those is the drawing of punnett squares in Drosophila genetics.
I wrote a little software (accessible here: silicocross.molecular.ch), that does basically that: drawing punnett squares.
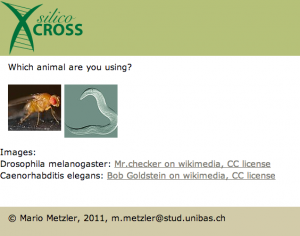
When you access the software you are asked how many chromosomes you want to track:
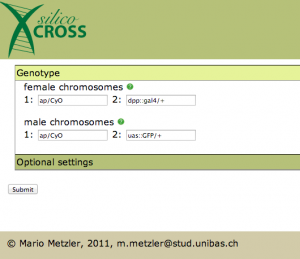
Let’s say we have a quite easy cross: on the second chromosome we have a mutant gene, that we want to have homozygous, and on the third we want to have a gal4 driver and a uas:GFP reporter.
So the number of chromosomes we want to track is two.
At this point you are asked for the model organism you are using, this is only relevant for the balancer chromosomes you are using. As at the moment I’m working only with flies, I suppose this will work better for basic tasks.
After you choose your favorite pet, it’s time to add it’s genotype:
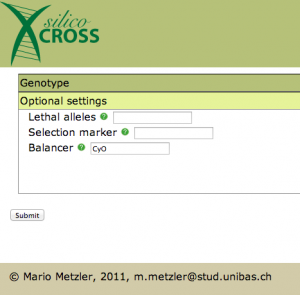
There is just one thing left to do now: we have to specify the balancer chromosome in Optional settings:
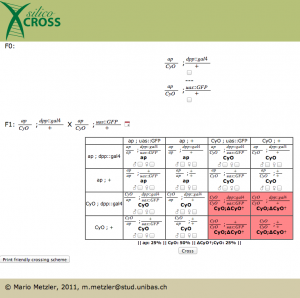
After pressing the submit button, the punnett square appears. At this point one could choose two genotypes for further crossing, this is not necessary in our case. Notice how death fly genotypes are highlighted in red. Sadly the gal4 system is not implemented yet, so in the phenotype you don’t see the GFP popping out (this is definitely in the to do list).
The system is still in heavy development (whenever I have some spare time and motivation), but I still use it in daily work when setting up crosses to have a nice graphic to glue in my fly-notebook or for teaching reasons. Hope it will be useful for the community as well.
At the moment I’m rewriting the core in python, as in my opinion this will give a less messed up code compared to the actual PHP core.
If you find bugs, have suggestions or else, don’t hesitate to contact me.



 (5 votes)
(5 votes)
Punnett squares are somewhat complex to use during daily lab life. Have you ever thought about a programm for the curly bracket scheme?
Dear Andreas,
This is indeed right.
I already kind of implemented that (it’s not working correctly yet, and it’s one of the reasons I’m rewriting the code).
In the punnett square view, after you finished your crossings, you can click on the “pretty print” button on the bottom left and you have the curly bracket view.
Best wishes
Mario
the accessible link is not working