Studying genealogy in cell clones
Posted by Fernando Garcia-Moreno, on 18 March 2014
A new method to study the beauty and relevance of cellular lineage
The origin of living beings has always interested, intrigued and fascinated curious researchers during the history of mankind. In the XIXth century, along with the flourishing Cell Theory and its “all living cells arise from pre-existing cells by division” the discipline of developmental biology was born. This new research area tried to understand the mechanisms by which a founder cell divides a limited number of times to give rise to a new organism. Gaining knowledge of cell lineage from progenitors in different tissues of the body became the “Holy Grail” of developmental biology. Lineage during embryonic development encloses all relevant biological information about formation and differentiation of the tissues and organisms under normal conditions, and it can explain a multitude of pathological conditions.
Unfortunately the cells of adult individuals don’t reveal much about their embryological past and therefore cell lineage is difficult to analyse without experimental intrusion. Cells, not just the ones that live in UK, don’t possess ID cards that contain their family names and their birthdates.
We considered these issues so important that we invested the last few years to establish a new labelling method named CLoNe for the study of cell lineages that can be used virtually in all types of tissues and amniote species (Garcia Moreno et al., 2014). In order to understand how it works I will make use of an analogy among the study of cell lineages and genealogy, the study of familiar lineages.
As mentioned above, adult cells don’t easily show information about their birthdate, birthplace, lineage relationships, such as surnames do, but some methodologies do actually allow distinguishing some cells from others. We can insert exogenous DNA into embryonic cells by means of viral particle vectors or through transfection by electroporation (a well-extended method in developmental neurobiology in which a low electric current transiently opens cell pores and guide and introduce through them DNA vectors previously injected in the tissue). This DNA usually carries a reporter gene, typically the easily detectable fluorescent protein GFP, which we could consider as the surname in the ID card of the cell. When a transfected cell expressing GFP divides, the daughter cells acquire, or should we say inherit, the expression of GFP. Therefore, we could define all GFP-expressing cells as the heir of a common cell lineage, couldn’t we?
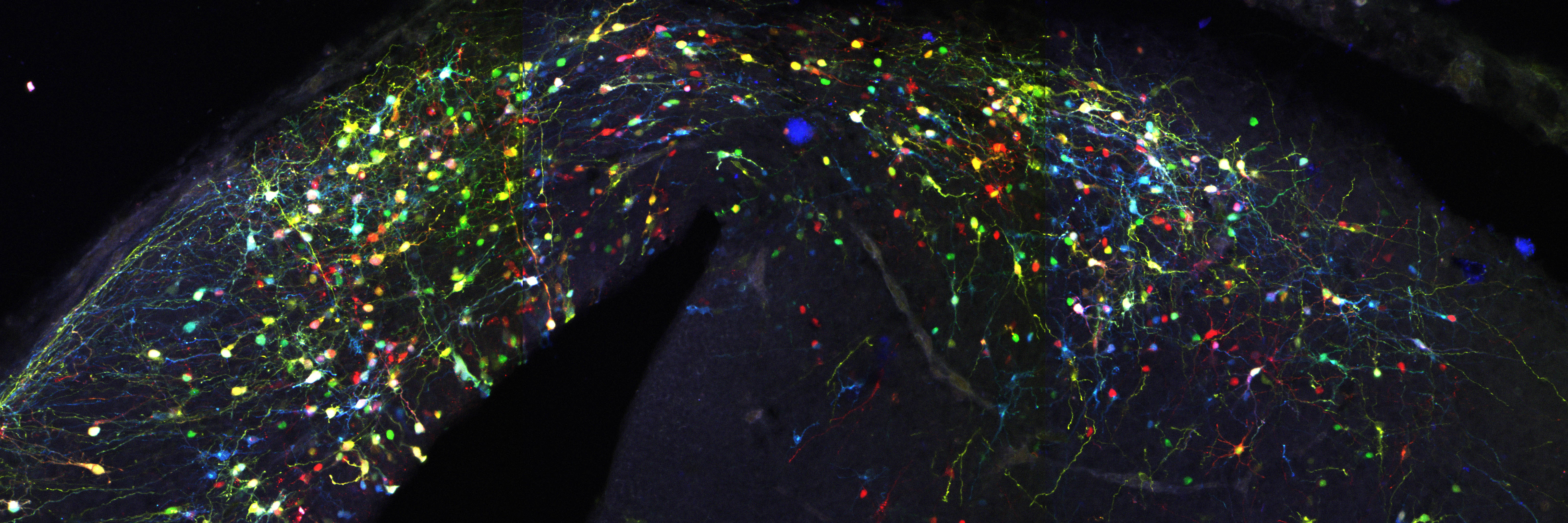
Actually not, since this simple system entails two major limitations. First that a single reporter gene doesn’t generate variability of surnames that would be enough to reliably discriminate among the billions of cells comprising a whole organism, e.g. someone is called Jones in Wales, or Molnár in Hungary. If we were all called Smith in UK, or García in Spain, it would be impossible for a genealogist to reveal the familiar origins and relationships of any of us. In our method CLoNe, inspired by the multi-coloured fluorescent Brainbow mice (Livet et al., 2007), we tackled this weakness by transfecting numerous distinct reporter genes, a total of twelve different fluorescent proteins. These 12 proteins are capable of generating thousands of different combinations on the basis of combinatorial hue, subcellular expression pattern of the fluorophores and intensity of the fluorescence (Fig. 1, observe the variability of fluorescent hues of the transfected cells in the chick brain). This extension in the number of reporter genes and their countless permutations is able to generate a vast amount of distinguishable surnames. Now, in addition to the Smiths we can distinguish the Jones and the Clayworths, or even the multiple-barrelled names (e.g. Smith-Jones-Clayworths–Howshams) that are truly unique. This is an idea that has been utilised in Spain for centuries (e.g. García-Moreno, or Manuel-Duarte-de Bendito) and now driving Pubmed Citations into a much more secure grounds.
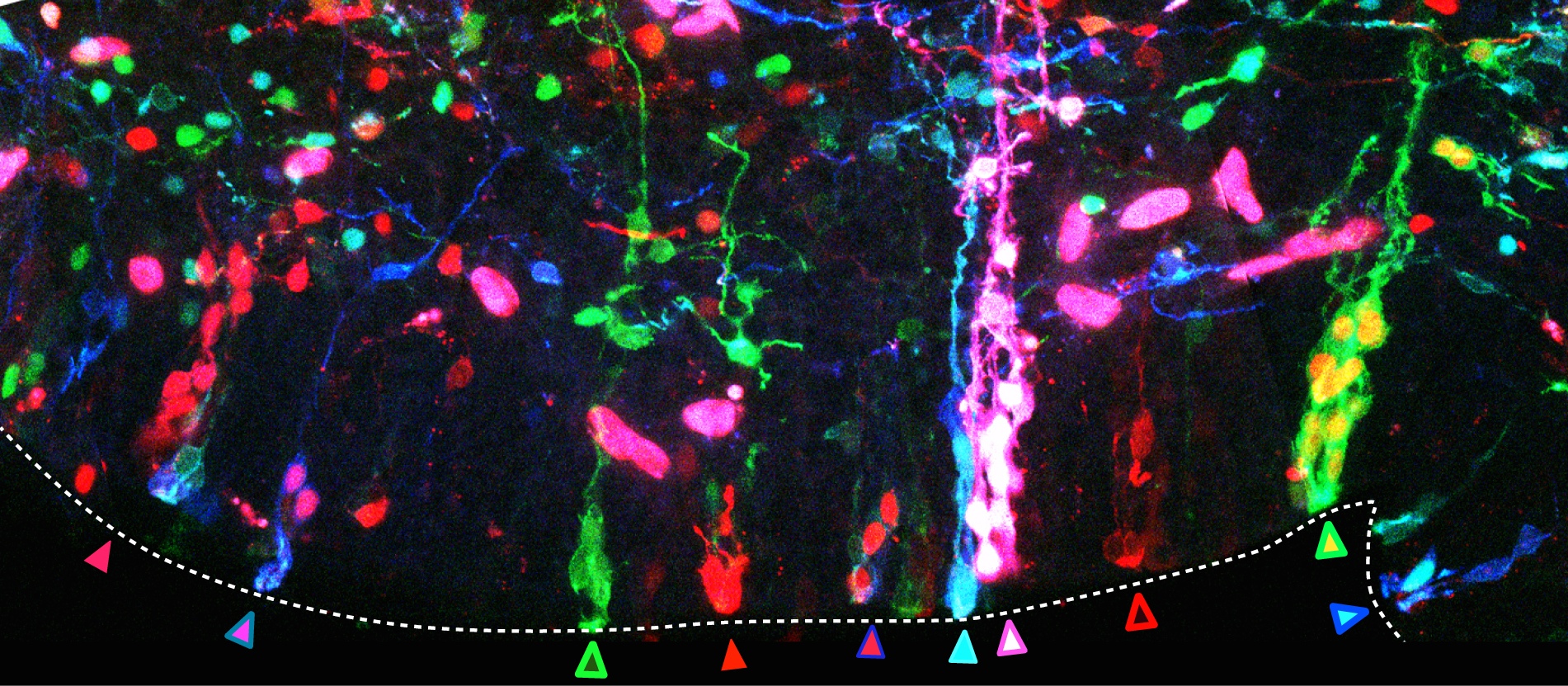
The second important drawback of the transfection by electroporation method implies the dilution of the fluorescent proteins till they are finally lost by successive cellular divisions (e.g. in Spain the family name is changing as you go down on generations, rather than sticking to the same multi-barrelled names). The transfected DNA constructs remain episomal, in the cellular cytoplasm, and distribute to the daughter cells during mitosis. Due to the finite number of DNA copies transfected, daughter cells inherit fewer and fewer DNA copies after each cell cycle and after a certain and unknown number of divisions, they don’t inherit the mark anymore. As a consequence, cells generated after the loss of the reporter gene are indistinguishable from non-transfected cells. So an unspecified portion of the cell lineage would be erroneously considered not belonging to the lineage. Imagine that after several generations the Smiths or the Clayworths forget their surnames. Our imaginary genealogist wouldn’t be able to relate individuals from different generations. In our method, influenced by Star Track method for labelling astrocytic clones (García-Marqués et al., 2013), we recombined the reporter genes into the nuclear genome of the transfected cells by means of the piggyBac transposon (Ding et al, 2005). Transposons are genetic elements that can jump to different locations within the genome. In our case, a random number and combination of the various reporter genes jump from the episomal constructs into the host genome of the transfected progenitor cell. During cell cycle, DNA synthesis duplicates the combination of reporter genes present in the genome and daughter cells inherit identical copies of the cellular surname, the combinatorial fluorescent expression (Fig. 2, progenitor cells at the germinal zone of the chick brain; chains of cells derived from the same stem cell share a common colour palette of fluorescent proteins). Back to genealogy (or should I say gene analogy?), our transposition system perpetuates the Clayworth surname in the family to all descendants; every new generation will always be called Clayworth. However, do we know that all the Clayworths are related and descend from the same original Clayworth?
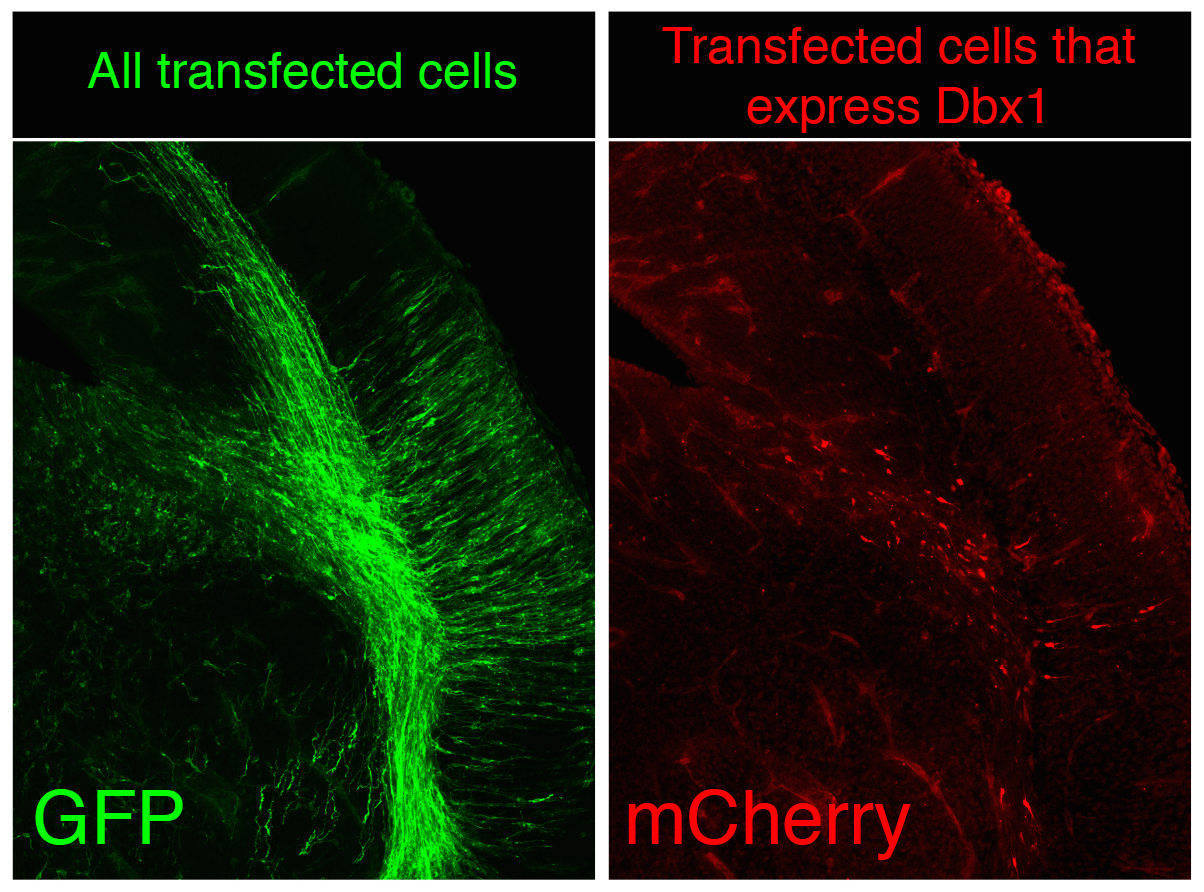
Although random integration by piggyBac is very capable to create a substantial number of surnames and though these are perpetuated in the genome the problem is not completely solved, since our ambitious genealogist doesn’t study a small town, but the biggest of the largest capitals, several magnitude orders over London, Mumbai or Mexico DF. There are millions of cells in the nervous system of a mouse, not to speak of the billions of cells in the human body. The countless combinations of surnames are simply not enough when considering billions of cells at once because the same random surname could occur independently in several progenitors during transposition. To avoid it, we apply two complementary strategies in CLoNe. First we reduce the number of transfected cells through the dilution of the Cre recombinase-expressing construct (which is responsible for the activation of the fluorescent labelling). Therefore in our particular cellular London, a small proportion of citizens do have a surname while a vast majority of the population doesn’t have a surname, so we can claim two individuals named Clayworth as relatives. Additionally, just as every genealogist does when refining the study to medieval kings, with CLoNe we decide a priori (before transfection) what cellular population we study the lineage in. During embryonic development stem cells express a variety of genes in order to perform their proliferative and regional identification functions. These genes are eventually very specific and identify a given population of progenitor cells. By associating the expression of Cre, and therefore the appearance of cellular surnames, to the expression of these identitary genes, CLoNe allows us to provide surnames to only our population of interest (Fig. 3, in the example we chose to selectively label in red cells expressing the transcription factor Dbx1). Remarkably this could be the main distinctive feature of CLoNe, the previous choice of the progenitor to be studied, instead of the a posteriori deduction of the labelled lineage. Ending the analogy, thanks to the employment of regulatory sequences of identitary genes, with CLoNe we can target and refine our study to the genealogy of the Clayworths from Notting Hill, or the Smiths from Kensington. And since we generate a great number of surnames we can always study the lineage of multiple cellular clones in the same piece of tissue (of multiple families in the same neighbourhood). The Clayworths, the Smiths, the Howshams AND the Skywalkers from Kensington can be studied at the same time, even though they are next-door neighbours. This cannot be achieved by other commonly used ways, such as viral methods, that only employ a single reporter gene.
CLoNe has been designed for a multitude of tissues and species, and has been tested in the nervous system, muscular and epithelial tissues of chick and mouse embryos. Its use can be extended to a large variety of experimental paradigms therefore it could be of interest to a variety of developmental biologists. By means of this new method we can generalise the study of cell lineage as always interesting, intriguing and fascinating. Not just biology or genealogy, in many aspects and disciplines the knowledge of lineages and kinships plays a crucial role, for otherwise Star Wars would have been only another space war movie.
References
Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y. and Xu, T. (2005). Efficient Transposition of the piggyBac (PB) Transposon in Mammalian Cells and Mice. Cell 122, 473–483.
García-Marqués, J. and López-Mascaraque, L. (2013). Clonal identity determines astrocyte cortical heterogeneity. Cereb. Cortex 23, 1463-1472.
García-Moreno, F., Vasistha, N., Begbie, J. And Molnár, Z. (2014). CLoNe is a new method to target single progenitors and study their progeny in mouse and chick. Development 2014 141:1589-1598; doi:10.1242/dev.105254.
Livet, J., Weissman, T. A., Kang, H., Draft, R. W., Lu, J., Bennis, R. A., Sanes, J. R. and Lichtman, J. W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62.



 (10 votes)
(10 votes)