In Development this week (Vol. 139, Issue 17)
Posted by Seema Grewal, on 7 August 2012
Here are the highlights from the current issue of Development:
eIF4E-3 puts a cap on spermatogenesis
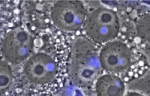 Gene expression is translationally regulated during many developmental processes. Translation is mainly controlled at the initiation step, which involves recognition of the mRNA 5′ cap structure by the eukaryotic initiation factor 4E (eIF4E). Eukaryotic genomes often encode several eIF4E paralogues but their biological relevance is largely unknown. Here (p. 3211), Paul Lasko and co-workers report that Drosophila eIF4E-3, one of eight fly eIF4E cognates, is essential for spermatogenesis. The researchers show that eIF4E-3 is a testis-specific protein and that male flies lacking eIF4E-3 are sterile. eIF4E-3 is required for meiotic chromosome segregation and cytokinesis, they report, and for nuclear shaping and sperm individualisation. The researchers also show that eIF4E-3 physically interacts with other components of the cap-binding complex. Furthermore, many proteins are expressed at different levels in wild-type and eIF4E-3 mutant testes, suggesting that eIF4E-3 has widespread effects on translation. These results add to the evidence that alternative forms of eIF4E add complexity to the control of gene expression during eukaryotic development.
Gene expression is translationally regulated during many developmental processes. Translation is mainly controlled at the initiation step, which involves recognition of the mRNA 5′ cap structure by the eukaryotic initiation factor 4E (eIF4E). Eukaryotic genomes often encode several eIF4E paralogues but their biological relevance is largely unknown. Here (p. 3211), Paul Lasko and co-workers report that Drosophila eIF4E-3, one of eight fly eIF4E cognates, is essential for spermatogenesis. The researchers show that eIF4E-3 is a testis-specific protein and that male flies lacking eIF4E-3 are sterile. eIF4E-3 is required for meiotic chromosome segregation and cytokinesis, they report, and for nuclear shaping and sperm individualisation. The researchers also show that eIF4E-3 physically interacts with other components of the cap-binding complex. Furthermore, many proteins are expressed at different levels in wild-type and eIF4E-3 mutant testes, suggesting that eIF4E-3 has widespread effects on translation. These results add to the evidence that alternative forms of eIF4E add complexity to the control of gene expression during eukaryotic development.
SnoN regulates mammary alveologenesis and lactation
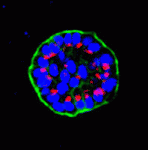 Mammary epithelial cells undergo structural and functional differentiation at late pregnancy and parturition to initiate milk secretion. TGF-β and prolactin signalling act antagonistically to regulate this process but what coordinates these pathways? On p. 3147, Kunxin Luo and colleagues report that SnoN, a member of the Ski family of pro-oncogenic and anti-oncogenic proteins, regulates both TGF-β and prolactin signalling to control alveologenesis and lactation in mice. The researchers show that the expression of SnoN, a negative regulator of TGF-β signalling, is induced at late pregnancy through the coordinated actions of TGF-β and prolactin. Heightened SnoN expression, they report, represses TGF-β signalling, which relieves TGF-β inhibition of the prolactin pathway. SnoN also directly promotes prolactin signalling by stabilising Stat5, a mediator of prolactin signalling. Consistent with these results, alveologenesis and lactogenesis are severely disrupted in SnoN–/– mice and mammary epithelial cells from these mice fail to undergo proper morphogenesis in vitro unless rescued by active Stat5 expression. Together, these results identify a new role for SnoN in the regulation of lactation.
Mammary epithelial cells undergo structural and functional differentiation at late pregnancy and parturition to initiate milk secretion. TGF-β and prolactin signalling act antagonistically to regulate this process but what coordinates these pathways? On p. 3147, Kunxin Luo and colleagues report that SnoN, a member of the Ski family of pro-oncogenic and anti-oncogenic proteins, regulates both TGF-β and prolactin signalling to control alveologenesis and lactation in mice. The researchers show that the expression of SnoN, a negative regulator of TGF-β signalling, is induced at late pregnancy through the coordinated actions of TGF-β and prolactin. Heightened SnoN expression, they report, represses TGF-β signalling, which relieves TGF-β inhibition of the prolactin pathway. SnoN also directly promotes prolactin signalling by stabilising Stat5, a mediator of prolactin signalling. Consistent with these results, alveologenesis and lactogenesis are severely disrupted in SnoN–/– mice and mammary epithelial cells from these mice fail to undergo proper morphogenesis in vitro unless rescued by active Stat5 expression. Together, these results identify a new role for SnoN in the regulation of lactation.
Co-operative neuronal migration
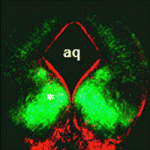 During the development of the central nervous system, neurons and/or neuronal precursors travel along diverse routes from the ventricular zones of the developing brain and integrate into specific brain circuits. Neuronal migration has been extensively studied in the forebrain but little is known about this key developmental event in the embryonic midbrain (mesencephalon). On p. 3136, Kwang-Soo Kim, Anju Vasudevan and co-workers remedy this situation by studying the migration of dopaminergic (DA) and GABAergic (GABA) neurons in the mouse mesencephalon. They show that DA and GABA neurons follow similar paths to the ventral mesencephalon (VM) in a temporally sequential manner. Interestingly, they report that in Pitx3-deficient (aphakia) mice, which have a defective DA neuron architecture, DA neuron migration is abnormal, stalled DA progenitors fail to reach the VM and GABA neurons also fail to migrate to the VM. These results suggest that pre-existing DA neurons modulate the migration of GABA neurons, thereby providing new insights into neuronal migration and the establishment of brain connectivity during mesencephalon development.
During the development of the central nervous system, neurons and/or neuronal precursors travel along diverse routes from the ventricular zones of the developing brain and integrate into specific brain circuits. Neuronal migration has been extensively studied in the forebrain but little is known about this key developmental event in the embryonic midbrain (mesencephalon). On p. 3136, Kwang-Soo Kim, Anju Vasudevan and co-workers remedy this situation by studying the migration of dopaminergic (DA) and GABAergic (GABA) neurons in the mouse mesencephalon. They show that DA and GABA neurons follow similar paths to the ventral mesencephalon (VM) in a temporally sequential manner. Interestingly, they report that in Pitx3-deficient (aphakia) mice, which have a defective DA neuron architecture, DA neuron migration is abnormal, stalled DA progenitors fail to reach the VM and GABA neurons also fail to migrate to the VM. These results suggest that pre-existing DA neurons modulate the migration of GABA neurons, thereby providing new insights into neuronal migration and the establishment of brain connectivity during mesencephalon development.
Out on a limb: HoxD chromatin topology
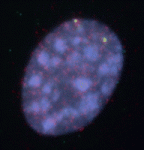 Anterior-posterior patterning of both the primary embryonic axis and the secondary body axis (limbs and digits) in mammals requires regulated Hox expression. Polycomb-mediated changes in chromatin structure control Hox expression during the first patterning event but are they also involved in the second? Here (p. 3157), Robert Hill, Wendy Bickmore and colleagues analyse the chromatin topology of the HoxD gene cluster in immortalised cell lines derived from posterior and anterior regions of distal E10.5 mouse limbs and in dissected E10.5 limb buds. They report that there is a loss of polycomb-catalysed histone methylation and a chromatin decompaction over HoxD in the distal posterior, compared with the anterior, limb. Moreover, the global control region spatially localises with the 5′ HoxD genomic region specifically in the distal posterior limb, a result that is consistent with chromatin looping between this long-range enhancer and its target genes. Thus, the researchers conclude, the development of the mammalian secondary body axis involves anterior-posterior differences in chromatin compaction and looping.
Anterior-posterior patterning of both the primary embryonic axis and the secondary body axis (limbs and digits) in mammals requires regulated Hox expression. Polycomb-mediated changes in chromatin structure control Hox expression during the first patterning event but are they also involved in the second? Here (p. 3157), Robert Hill, Wendy Bickmore and colleagues analyse the chromatin topology of the HoxD gene cluster in immortalised cell lines derived from posterior and anterior regions of distal E10.5 mouse limbs and in dissected E10.5 limb buds. They report that there is a loss of polycomb-catalysed histone methylation and a chromatin decompaction over HoxD in the distal posterior, compared with the anterior, limb. Moreover, the global control region spatially localises with the 5′ HoxD genomic region specifically in the distal posterior limb, a result that is consistent with chromatin looping between this long-range enhancer and its target genes. Thus, the researchers conclude, the development of the mammalian secondary body axis involves anterior-posterior differences in chromatin compaction and looping.
Developmental roles for ribosomal biogenesis genes
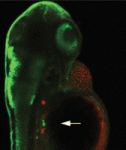 Mutations in the human Shwachman-Bodian-Diamond syndrome (SBDS) gene, which functions during maturation of the large 60S ribosomal subunit, cause a disorder characterised by exocrine pancreatic insufficiency, chronic neutropenia and skeletal defects. Steven Leach and colleagues have now refined a zebrafish model of this ‘ribosomopathy’ (see p. 3232). Knockdown of the zebrafish sbds orthologue, they report, fully recapitulates the developmental abnormalities of the human syndrome but, interestingly, unlike in other ribosomopathies, loss of p53 does not rescue these developmental defects. The researchers show that impaired proliferation of pancreatic progenitor cells is the primary defect underlying the pancreatic phenotype and report that loss of sbds results in widespread changes in the expression of genes related to ribosome biogenesis, rRNA processing and translational initiation, including ribosomal protein L3 and pescadillo. Notably, inactivation of either of these genes also impairs expansion of pancreatic progenitor cells in a p53-independent manner. Together, these results suggest new p53-independent developmental roles for ribosomal biogenesis genes.
Mutations in the human Shwachman-Bodian-Diamond syndrome (SBDS) gene, which functions during maturation of the large 60S ribosomal subunit, cause a disorder characterised by exocrine pancreatic insufficiency, chronic neutropenia and skeletal defects. Steven Leach and colleagues have now refined a zebrafish model of this ‘ribosomopathy’ (see p. 3232). Knockdown of the zebrafish sbds orthologue, they report, fully recapitulates the developmental abnormalities of the human syndrome but, interestingly, unlike in other ribosomopathies, loss of p53 does not rescue these developmental defects. The researchers show that impaired proliferation of pancreatic progenitor cells is the primary defect underlying the pancreatic phenotype and report that loss of sbds results in widespread changes in the expression of genes related to ribosome biogenesis, rRNA processing and translational initiation, including ribosomal protein L3 and pescadillo. Notably, inactivation of either of these genes also impairs expansion of pancreatic progenitor cells in a p53-independent manner. Together, these results suggest new p53-independent developmental roles for ribosomal biogenesis genes.
Mapping the mouse embryo
 The sequence and location of every gene in the human genome is now known but our understanding of the relationships between human genotypes and phenotypes is in its infancy. To better understand the role of every gene in the development of an individual, the International Mouse Phenotyping Consortium aims to phenotype targeted gene knockout mice throughout the genome (∼23,000 genes). Because many of these mice will be embryonic lethal, methods for phenotyping mouse embryos are needed. Michael Wong and colleagues are developing such an approach and, on p. 3248, they present a new three-dimensional atlas of the mouse embryo. To produce their atlas, the researchers combined micro-computed tomography images of 35 E15.5 mouse embryos into an average image using automated image registration software, and then manually segmented 48 anatomical structures. This atlas establishes baseline anatomical phenotypic measurements against which mutant mouse phenotypes can be assessed; in the future, a mutant embryo image can be registered to the atlas and its organ volumes calculated automatically.
The sequence and location of every gene in the human genome is now known but our understanding of the relationships between human genotypes and phenotypes is in its infancy. To better understand the role of every gene in the development of an individual, the International Mouse Phenotyping Consortium aims to phenotype targeted gene knockout mice throughout the genome (∼23,000 genes). Because many of these mice will be embryonic lethal, methods for phenotyping mouse embryos are needed. Michael Wong and colleagues are developing such an approach and, on p. 3248, they present a new three-dimensional atlas of the mouse embryo. To produce their atlas, the researchers combined micro-computed tomography images of 35 E15.5 mouse embryos into an average image using automated image registration software, and then manually segmented 48 anatomical structures. This atlas establishes baseline anatomical phenotypic measurements against which mutant mouse phenotypes can be assessed; in the future, a mutant embryo image can be registered to the atlas and its organ volumes calculated automatically.
Plus…
Making waves: the rise and fall and rise of quantitative developmental biology
 The tenth annual RIKEN Center for Developmental Biology symposium ‘Quantitative Developmental Biology’ held in March 2012 covered a range of topics. As reviewed by Davidson and Baum, the studies presented at the meeting shared a common theme in which a combination of physical theory, quantitative analysis and experiment was used to understand a specific cellular process in development. See the Meeting Review on p. 3065
The tenth annual RIKEN Center for Developmental Biology symposium ‘Quantitative Developmental Biology’ held in March 2012 covered a range of topics. As reviewed by Davidson and Baum, the studies presented at the meeting shared a common theme in which a combination of physical theory, quantitative analysis and experiment was used to understand a specific cellular process in development. See the Meeting Review on p. 3065
A computational image analysis glossary for biologists
 Meyerowitz and colleagues present a glossary of image analysis terms to aid biologists and discuss the importance of robust image analysis in developmental studies.
Meyerowitz and colleagues present a glossary of image analysis terms to aid biologists and discuss the importance of robust image analysis in developmental studies.
See the Primer article on p. 3071
Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies
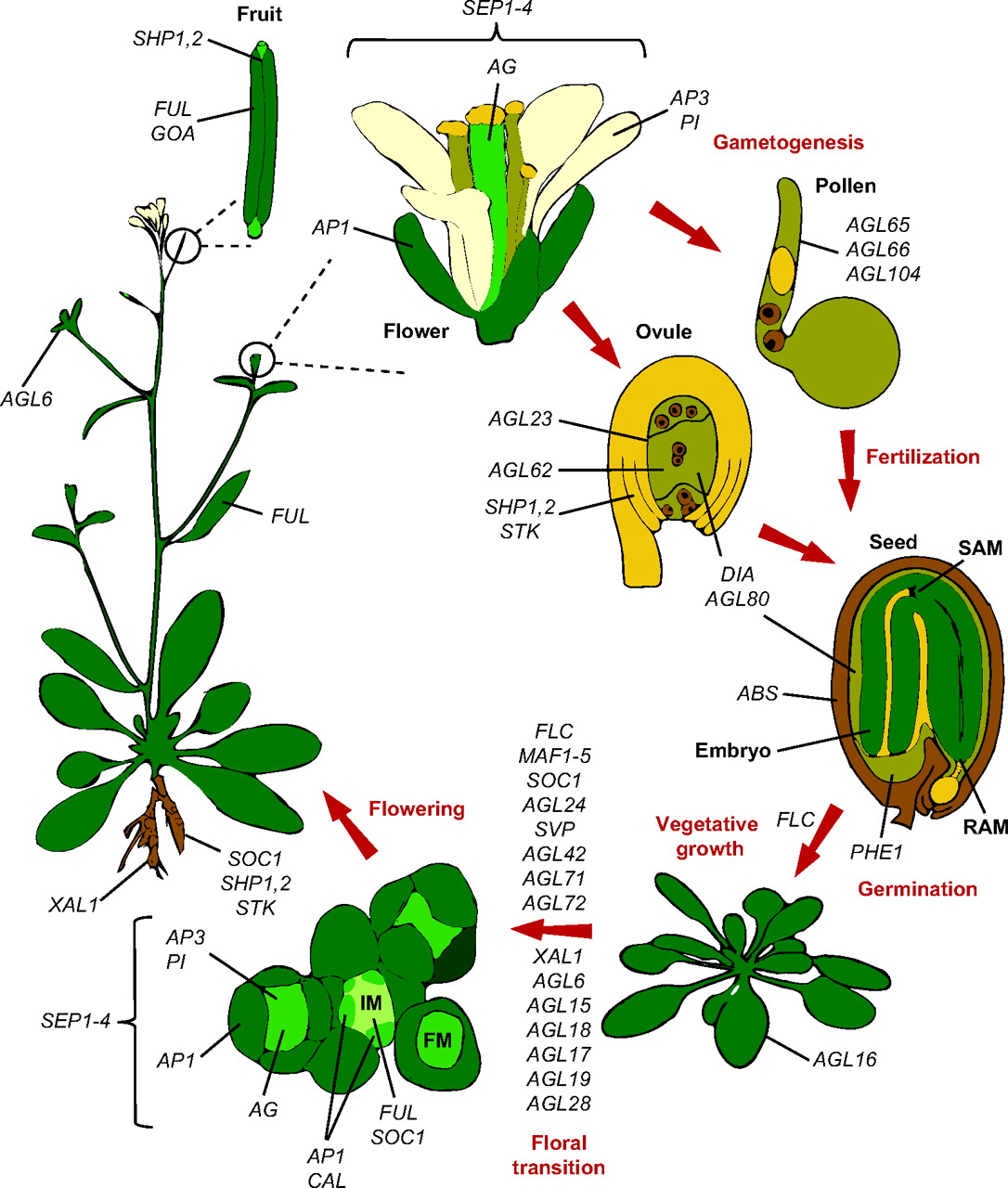 Members of the MADS-box transcription factor family play essential roles in almost every developmental process in plants. Kaufmann and colleagues review recent findings on MADS-box gene functions in Arabidopsis and discuss the evolutionary history and functional diversification of this gene family in plants.
Members of the MADS-box transcription factor family play essential roles in almost every developmental process in plants. Kaufmann and colleagues review recent findings on MADS-box gene functions in Arabidopsis and discuss the evolutionary history and functional diversification of this gene family in plants.
See the Review article on p. 3081


 (1 votes)
(1 votes)