Winding road to the cambial stem cells
Posted by Ari Pekka Mähönen, on 13 January 2025
In plants, the vascular cambium, a bifacial stem cell niche, drives wood formation by generating the xylem on one side and the phloem on the other. In this post, Ari Pekka Mähönen, Peter Etchells and Kirsten ten Tusscher tell the story behind their paper “Identification of cambium stem cell factors and their positioning mechanism”.
Ari Pekka Mähönen:
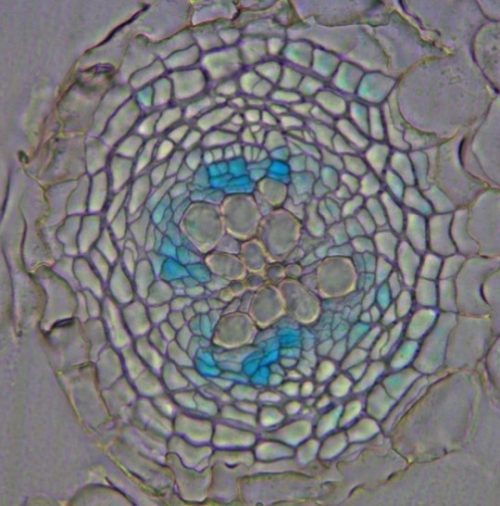
In the autumn of 2009, I returned from my post-doctoral period in Ben Scheres’ lab, located then in Utrecht. During my postdoc I was working on the roles of PLETHORA/AINTEGUMENTA-LIKE (PLT/AIL) transcription factors in stem cell regulation in the root meristem. Moving back to Finland my idea was to study whether any of these factors are expressed in Arabidopsis root cambium, the meristem I intended to study in my newly established research group. It was exciting to see that PLT5 showed very specific expression in the dividing cambial cells (Figure), however, further studies had to wait for several years due to my focus on finalizing ongoing projects. After obtaining funding for my research group, a PhD student, Gugan Eswaran, started to work on the project, and he discovered that ANT, PLT3 and PLT7 are also expressed in the cambium, suggesting genetic redundancy in cambium development. Unfortunately, ant single mutants and plt triple mutants showed only slightly reduced secondary growth. I was expecting a stronger phenotype from putative stem cell factors of cambium. In subsequent attempts, generation of the quadruple mutant failed due to unexplained lethality, and an artificial microRNA approach did not provide stronger phenotypes either. This was disappointing and thus Gugan started to focus more on his side projects. Then, rescue for the project came a few years later from a technical innovation. Xin Wang, a PhD student in my lab, invented an inducible genome editing system (IGE) for plants (Wang et al Nature Plants 2020). This IGE system was used to generate a conditional quadruple ant/plt mutant. Finally, this mutant showed severely reduced radial growth, something one would expect from loss of stem cell factors. This was great, but of course now we had a new question to answer – what regulates CAMBIUM-EXPRESSED AINTEGUMENTA-LIKE (CAIL) (the name we gave to ANT, PLT3, PLT5 and PLT7) expression in such a narrow, specific domain? I think, this was the point where you Peter contacted me, or was it even earlier than when we got the conditional quadruple mutant results?
Peter Etchells:
I got in touch while Gugan was making the IGE construct. I had been working on putting the pieces of PXY signalling together since joining Simon Turner’s lab in Manchester in 2007, and this continued when I moved to Siobhán Brady’s lab at UC Davis in 2013. Over that whole period, through my work and that of others in Hiroo Fukuda’s lab, a series of PXY-downstream targets, WOX4, WOX14, BES1, LBD4 and TMO6 were identified (Hirakawa et al Plant Cell 2012; Etchells et al Development 2013; Kondo et al Nature Comms 2015; Smit et al Plant Cell 2020). However, I was never satisfied that all the transcriptional targets of TDIF-PXY had been uncovered. PXY is homologous to CLAVATA1, which famously regulates the shoot apical meristem via regulation of the homeodomain transcription factor WUSCHEL (WUS). WOX4 and WOX14 are homologous to WUS, so they were a natural focus for investigation, but wox4 wox14 mutants only have a mild cambium phenotype. BES1, LBD4 and TMO6 are also only responsible for regulation of a subset of the pxy phenotypes. It seemed like we were missing something. The key was a transcriptomic experiment which demonstrated that CAIL genes were differentially expressed in both pxy and TDIF over-expression lines, performed just as I was transitioning out of Siobhán’s group to start my own lab in 2015. To test for a genetic interaction between the CAILs and TDIF-PXY, we crossed the TDIF over-expression line, which is characterised by ectopic cambium, to plt357 mutants. Although the plt357 line alonedid not have a cambium phenotype, it did suppress phenotypes associated with TDIF over-expression, which, combined with the CAIL expression patterns that Ari Pekka’s group had, demonstrated that the CAIL genes did have a cambium function and were likely controlled by TDIF-PXY. It was not long after that Gugan’s IGE line came through which sealed the deal. Still, the story was incomplete because the PXY expression domain is so broad relative to that of the CAILs.
Ari Pekka Mähönen:
So, now we knew that CAILs are regulated by the TDIF-PXY ligand-receptor pathway. However, we still did not know how come CAILs are only expressed in such a narrow region in stem cells, given that the PXY receptor expression domain spans from the stem cells into the xylem. A few researchers in my lab participated to hunt for the mechanism underlying this tight spatial control, and we indeed found a few regulatory feed-back mechanisms that could help excluding the CAILs from the xylem. On top of this, we wondered whether efficient sequestration of diffusing TDIF peptide by the PXY receptor could play a role in focusing CAIL signalling. With all the feedback regulation and a possible sequestration of TDIF, we were quite unsure which one of these mechanisms (or whether any of these mechanisms) could contribute to narrow CAIL expression in planta. Therefore, I contacted Kirsten ten Tusscher, a computational biologist, with whom I had collaborated before on addressing the role of PLT genes in root zonation (Mähönen, ten Tusscher et al. Nature 2014). I suggested that we could address these different scenarios in TDIF-PXY-CAIL signalling in a computational model.
Kirsten ten Tusscher:
As I had greatly enjoyed our previous collaboration, and questions on patterning are the bread and butter of computational biology, this was of course an offer I could not refuse. Luckily, a talented PhD student in my group, Jaap Rutten, was quite far already with the results for the main project of his PhD thesis and waiting for experimental data. Thus, it was not hard at all to convince him to broaden his horizon beyond the control of root meristem size that we were working on together with Sabrina Sabatini to the control of cambium patterning and positioning together with Ari Pekka and Peter. To investigate the importance of different feedback mechanisms as well as the potential of ligand sequestration in defining the narrow domain of CAIL cambium expression, we started building a model incorporating all the important molecular players and the regulatory interactions between them, using both new and previously published data. As a start we developed a model for a single cell and tested whether it could model xylem, phloem and cambium cells depending on the incoming signals. However, for non-modelers it often seems that if models are complex enough and you tweak parameters you can make them do anything you want. Therefore, it was important to show that the models’ capacity to simulate phloem, xylem or cambium forming cells was a very generic property of the modelled network architecture, not of precise parameter values or details. To achieve this Jaap performed a whopping 1,768,593,750 simulations to extensively test different model settings, occupying some of our computers for weeks, and show that overall model behaviour remained the same. In the process we could already confirm that some of the feedback uncovered by Ari Pekka’s team indeed limited cambium formation and promoted xylem formation. As a next step we could now move to a one-dimensional model of a strip of cells spanning from xylem to phloem and start testing the ligand-sequestration hypothesis. Key to this was to include an auxin-dependent PXY gradient starting from the xylem end of the tissue, and a TDIF gradient arising from the diffusion of TDIF from TDIF producing phloem cells into our model. With this in place, Jaap demonstrated that if binding of TDIF ligand to PXY receptors is sufficiently strong, at the tissue position where TDIF meets the first low levels of PXY receptors, TDIF is bound and effective diffusion is halted, preventing TDIF-PXY interaction further towards the xylem. Interestingly, this also explains why cambium stem cell patterning is robust under various cambium sizes: when the xylem and phloem are further apart, the TDIF will diffuse further before it reaches the first PXY receptors and until that time it diffuses freely ensuring it will still meet PXY receptors! However, an important issue remained: the regulatory feedback mechanism uncovered earlier could to some extent limit CAIL expression. So, to test which of these potential mechanisms occurs and/or is most important in planta, we tested in silico what would happen if we decreased PXY expression or elevated TDIF levels. Next these experiments were also performed in the lab, with lab outcomes matching the predictions made by the sequestration-based model. This finally enabled us to cement the importance of sequestration for defining the CAIL expression domain.
Ari Pekka Mähönen, Peter Etchells, Kirsten ten Tusscher:
So, now we could confidently say that sequestration of TDIF is the key to focusing the TDIF-PXY signalling and thus CAIL expression in a narrow domain to define the stem cells. The manuscript was submitted, and we got constructive comments from the reviewers, especially on providing more evidence for the sequestration mechanism. Xixi Zhang, a post doc in the Mähönen lab, had already earlier started to work on the generation of PXY reporter lines. She noticed, among other findings, that the translational reporter pPXY:PXY-YFP has a significantly sharper gradient within the cambium than the transcriptional reporter line pPXY:erYFP, indicating that the PXY-YFP fusion protein is more unstable in phloem-side cambium cells than in the cells on the xylem-side of the cambium. Since TDIF ligand originates from phloem, this suggest that the TDIF binding to PXY could make PXY-YFP unstable. Thus, regulation of TDIF-PXY stability could be the key mechanism for the sequestration, and this is what Xixi is studying now, as a follow up of the published work.
In the end, seeing this paper published was particularly satisfying, both because of the long journey it took to finalize it and because of the enjoyable collaboration we had while working together on this project.


 (3 votes)
(3 votes)