Behind the paper: “Recording morphogen signals reveals origins of gastruloid symmetry breaking”
Posted by Harry McNamara, on 17 January 2025
Stem cell models as laboratories to study self-organization
My road from physics to developmental biology began in a journal club during my PhD in Adam Cohen’s lab at Harvard. We were discussing the first reports from Madeline Lancaster, Jürgen Knoblich, and their co-authors(Lancaster et al., 2013) describing the self-organization of cerebral organoids from stem cells. I was instantly fascinated by the images they reported: while no one would mistake these in vitro structures for a real brain, they still showed remarkably complex patterns of gene expression and tissue morphologies. In the physics community, there is longstanding interest in self-organizing systems: patterns that do not follow an external blueprint, but rather emerge from feedbacks in interactions between a system’s components. These stem cell models demonstrated the power of biological self-organization, and offered simplified ‘physics laboratories’ for decoding how multicellular programs arise from basic interactions between cells.
After completing my PhD, I moved to the Lewis-Sigler Institute (LSI) at Princeton as an independent fellow to study stem cells and organoids. The scientific community at the LSI is deeply interdisciplinary and has a long tradition of excellence in both developmental biology and biological physics. Much of this success came from leveraging the fruit fly embryo (D. melanogaster) as a quantitative system for studying fundamental principles of pattern formation. Fly embryos are highly amenable to quantitative measurement and genetic perturbation, and they can be produced at scale to achieve statistical power. I hoped that stem cell models could offer similar advantages, while opening new questions in developmental biophysics.
Choosing a problem: symmetry breaking in the gastruloid
I arrived at Princeton in January 2021 – a challenging time to start a postdoc anywhere, but my transition was made easier by the collegial and collaborative nature of the Princeton community. I was broadly interested in stem cell self-organization, and I began by searching for a self-organizing program to ‘decode’. I found a collaborator and mentor in Jared Toettcher. Jared is a bioengineer and molecular biologist whose group (together with Stas Shvartsman’s group, also at Princeton) had done pioneering work applying ideas from signal processing to the role of the Erk signaling pathway in fly development. Recently, he and his student Evan Underhill had started studying a stem cell model called the gastruloid(van den Brink et al., 2014).
The gastruloid recapitulates aspects of gastrulation: specifically, the formation and morphology elongation of an anterior-posterior axis. Evan and Jared were studying how FGF signals produced in the posterior end of the gastruloid activated downstream Erk and Akt signals to control patterning and elongation. I became interested in an earlier aspect of gastruloid formation: how does the gastruloid break symmetry to establish a posterior pole in the first place?
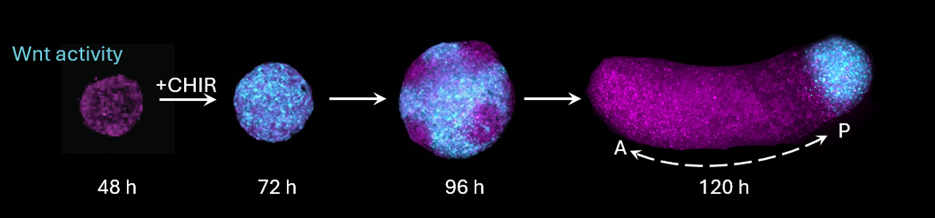
The mystery stems from the lack of explicit spatial cues which the gastruloid receives. In vivo, gastrulation is patterned by spatial cues provided by extra-embryonic tissues, including Wnt signaling molecules provided to the posterior epiblast. Gastruloid formation is triggered by activating Wnt signaling activity everywhere with the small molecule CHIRON-99201 (‘CHIR’). Gastruloids eventually form a polarized domain of Wnt activity in the posterior, just as the embryo does – but how does this local domain form when all cells receive the same stimulus?
Theories of self-organization and pattern formation offer candidate explanations. One possibility is that gastruloid polarization could be an example of Alan Turing’s reaction-diffusion theory of pattern formation(Turing, 1952): that is, Wnt-dependent feedback in the production of diffusing activators and inhibitors of Wnt signaling can amplify small differences and eventually spontaneously restrict activity to one local domain. Another possibility is that cells spatially rearrange to sort themselves into different domains as they change signaling levels. Discerning between these candidate models is extremely challenging: both could explain the patterns of Wnt signaling activity we observe during symmetry breaking and morphogenesis.

Recording signals to decode self-organization
Ideally, we could trace the history of signals that cells see and ask: when can we predict cell fates based on early signaling states? By measuring this kind of ‘fate information’, we could identify when the gastruloid breaks symmetry, and test predictions of mechanistic models. But this is also a very hard problem: cells communicate through many different signaling pathways, and optically tracking cells through divisions and migrations over several days of gastruloid morphogenesis would be extremely challenging.
To trace signaling histories of cells, we turned to synthetic biology. When I was in graduate school, I had followed work from Alex Schier’s group (then at Harvard MCB) developing new technologies for recording cells’ lineage within the genome(Farrell et al., 2018; McKenna et al., 2016). I realized that if this ‘molecular recording’ strategy were adapted to record signaling states (rather than lineage), it could offer us a tool to reconstruct signaling histories without solving the live-imaging cell tracking problem. (As an aside: many other groups have shared this same insight; signal recording is now a robust area of biotechnology development!)
Fortunately, Michelle Chan had just joined the LSI and Molecular Biology Department at Princeton. Michelle is an expert in lineage tracing and had done pioneering work building molecular lineage recorders in the mouse embryo(Chan et al., 2019). With her advice and guidance, we were able to weigh tradeoffs between different design strategies. After prototyping a few flavors, we settled on a recombinase-based strategy. Recombinases (the most famous of which is an enzyme called Cre) can make site-specific modifications at target sites: for example, excising a red fluorescent protein from the genome to permit expression of a green one. We adapted this classical lineage tracing strategy by embedding Cre within a genetic circuit that only expresses in the presence of two inputs: (1) signaling activity in a pathway of interest, and (2) a small molecule (doxycycline, or ‘dox’) used to gate a ‘listening window’ in which the circuit is queried. This logic allows us to relate the output of the Cre recording activity in a target signaling pathway within a known time window.
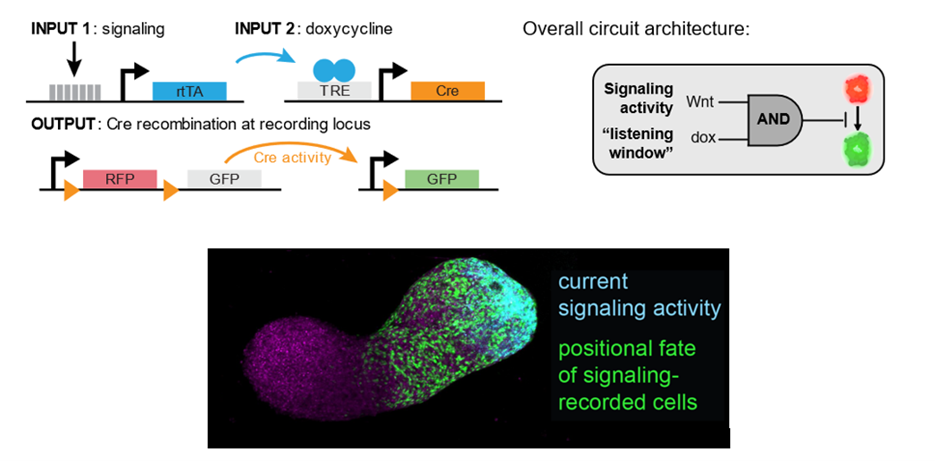
In many ways, this approach is quite old-school compared to other emerging technologies for signal recording. As designed, each recombinase only can record one bit of information in a single signaling pathway. But while our design sacrifices in information bandwidth, it gains exceptional sensitivity and fidelity: we could reliably resolve signaling states with high confidence within developmentally relevant temporal windows (3-6 hours). While this approach may not scale to the unbiased screening of all signaling pathways, in many developmental contexts we have a strong prior to focus on a few signals of outsized importance. We viewed this tradeoff of channel bandwidth for sensitivity and fidelity as favorable for our application.
As a practical matter, the main challenge in building these designs into cells was tuning the sensitivities of relevant components. There is a strong ‘Goldilocks principle’ at play: if signal recording is too sensitive, we may get ‘leaky’ background activity even in the absence of signaling activity or dox. But if it is not sensitive enough, then we may not be able to resolve signaling activity effectively. Ideally, the circuit sensitivities should be balanced to be just right.
One way to alter component sensitivities is via the genomic context around the integration site of our synthetic genes. We developed a cell engineering pipeline to screen libraries of randomly integrated parts and then select-out the candidates which landed in the Goldilocks zone. We ultimately identified high-performing cell lines which record each of three canonical signaling pathways that orchestrate gastrulation: Wnt, Nodal, and BMP. The latter two were crucially enabled by work from Ken Zaret’s lab, which validated a panel of pathway-specific sentinel promoters(Serup et al., 2012). This work is a wonderful resource for the community, and we are grateful that they shared constructs of for Nodal- and BMP-responsive elements (AR8 and IBRE4).
Putting theories of self-organization to the test
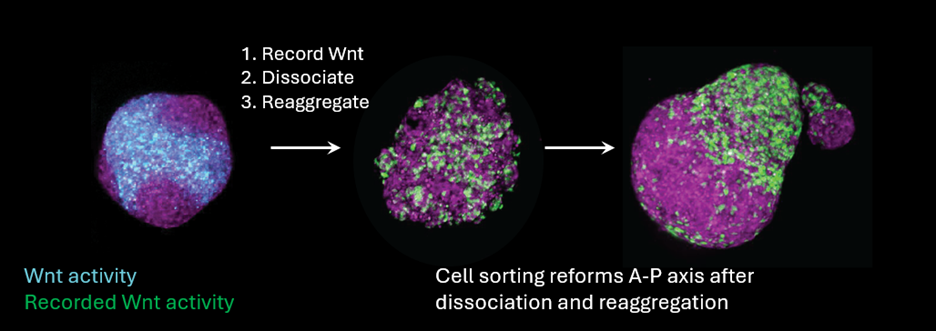
With our signal-recording cell lines in hand, we set out to systematically map when early Wnt, Nodal, and BMP signaling states predict future cell fates. I won’t belabor all of our results here – please read our paper for the complete story! But I will describe my favorite experiment. One surprising observation was that when we recorded Wnt activity during a ‘patchy’ state – that is, when signaling activity was locally correlated into domains, but not yet globally polarized – we could already predict future cell fates along the final A-P axis. It seemed that cell rearrangements (and *not* reaction-diffusion feedbacks) were sorting the patchy domains into a single pole. But we still weren’t completely certain of our interpretation – more complex reaction-diffusion models could in principle still be consistent with our data.
We put our model to the test by testing its predictions more explicitly. Under a strong cell-sorting model, we should be able to record Wnt states with our signal recorder, and then scramble cell positions by dissociating (with Trypsin) and then reaggregating to form new spheroids. We did the experiment, and the result was clear as day – the red and green cells managed to find each other and phase-separate into different domains! We also incidentally observed that reaggregating sometimes made smaller ‘satellite’ gastruloids that sorted into a scaled-down pattern (which we then confirmed by deliberately making smaller reaggregate gastruloids with a flow cytometer). Scaling follows nicely from a cell sorting model: as long as you have the right balance of cell types, sorting gives you scaling for free.
Our synthetic biology approach provided us with an integrated description of how the gastruloid self-organizes and allowed us to interrogate fundamental theories of pattern formation. Many interesting questions remain: for example, what are the physics of gastruloid cell sorting? Are there even earlier physical or biochemical cues which contribute to symmetry breaking? I am also excited about opportunities to use synthetic biology to control signaling states in the gastruloid, and to use this synthetic biology toolbox to decode self-organization in other organoid and embryoid models. In January 2025, I moved to Yale to start a new research group to chase these questions. Please reach out if you are interested in joining the adventure!
References
Chan, M. M., Smith, Z. D., Grosswendt, S., Kretzmer, H., Norman, T. M., Adamson, B., Jost, M., Quinn, J. J., Yang, D., Jones, M. G., et al. (2019). Molecular recording of mammalian embryogenesis. Nature 570, 77–82.
Farrell, J. A., Wang, Y., Riesenfeld, S. J., Shekhar, K., Regev, A. and Schier, A. F. (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131.
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., Homfray, T., Penninger, J. M., Jackson, A. P. and Knoblich, J. A. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379.
McKenna, A., Findlay, G. M., Gagnon, J. A., Horwitz, M. S., Schier, A. F. and Shendure, J. (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907.
Serup, P., Gustavsen, C., Klein, T., Potter, L. A., Lin, R., Mullapudi, N., Wandzioch, E., Hines, A., Davis, A., Bruun, C., et al. (2012). Partial promoter substitutions generating transcriptional sentinels of diverse signaling pathways in embryonic stem cells and mice. Disease Models & Mechanisms 5, 956–966.
Turing, A. M. (1952). The Chemical Basis of Morphogenesis. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 237, 37–72.
van den Brink, S. C., Baillie-Johnson, P., Balayo, T., Hadjantonakis, A.-K., Nowotschin, S., Turner, D. A. and Martinez Arias, A. (2014). Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141, 4231–4242.


 (1 votes)
(1 votes)