A short rant on the present and future of developmental biology
Posted by Jonas Hartmann, on 11 September 2024
Observing a cluster of migrating cells or a developing embryo through the lens of a microscope can be a visceral experience; one is struck by the ephemeral beauty, layered complexity, and alien intelligence displayed by such specimens. For those who seek a scientific understanding of these striking phenomena, it is also a humbling experience. There are so many moving parts here, so many subsystems within subsystems, so much noise, so much nonlinearity, so much contingency… how could we possibly hope to capture this in the simple yet powerful models that make scientific explanations so satisfying and useful?
I’ve been grappling with this question ever since my undergrad, and as anyone who does so, I have found plenty of reasons to be pessimistic about it.
Though we have extensive knowledge of the molecular machines that form the building blocks of biological systems, putting this giant puzzle together from the bottom up seems an impossibly complicated task. Instead, the field’s still-dominant approach is to link particular perturbations to particular outcomes, usually by lifting out a handful of mechanisms or genes from the broader system, drawing arrows between them, and calling the result a pathway. But whilst the models this produces are of an appealing simplicity, they lack power; they often fail to explain or predict anything outside the narrow set of conditions and observations that were considered in the original study. At least we are indisputably making progress in developing new tools to collect more and better data, ever more quickly, ever more precisely… but alas, this progress is closely shadowed by the realization that it can only take us so far; more data does not yield more understanding if we don’t know how to ask the right questions.
With these problems permeating the field, it comes as no surprise that there is a measure of discontent in the community. Some argue that we have an attitude problem [1]; perhaps young researchers spend too much time on twitter and not enough time in the library? Others contend that we have an image problem [2]; perhaps we should be spending more time on twitter, reassuring each other and the wider public that our field remains essential – or even that it has recently entered, as some would have it [3], a “new golden age”? Like so many developmental biology papers, these viewpoints may not be entirely wrong, but they’re also not particularly compelling.
I’d like for us to entertain the possibility that we are in fact facing a science problem. That our progress is not bottlenecked by modern attitudes or public misperceptions, but by the profound intellectual challenge of finding new and better ways of thinking about the spectacles that play out under our microscopes. I’d like to take seriously the above reasons for pessimism and treat them as real scientific challenges for us to tackle and overcome. If the molecular details are intractable, we should search for new and better systems-level abstractions to subsume them. If the current standard of mechanistic explanation is inadequate, we should look to build new and better conceptual frameworks that set a higher standard. If it is hard to distill meaning from the deluge of high-throughput data, we should aim to develop new and better models that yield strong inductive priors for big-data analysis.
This is much easier said than done, of course, but in grappling with these issues I have also come across a few reasons for optimism!
Looking back in history, the challenge faced by Darwin and his contemporaries in seeking to unify the diversity of living organisms must have seemed no less daunting than our current predicament, yet they persevered and emerged with an entirely new understanding of life. Returning to modern times, a new theory of cell types established about a decade ago shows brilliantly that deep conceptual progress is possible even today [4]. And not only that; it also shows that such progress really does have the impact we would hope to see! For one, it has inspired new ways of analyzing and interpreting transcriptomics data (see e.g. [5]). For another, I have personally witnessed how much more productive the discourse on cell types can be within a group of researchers who know this theory (even if they don’t all fully endorse it) compared to a group who do not. These and other inspirational observations are always in the back of my mind as I explore my own ideas for tackling the field’s fundamental problems.
One such idea is the Core & Periphery (C&P) hypothesis, which was published last week [6] and serves as the occasion for this post.
The C&P idea originated from discussions between first author Elisa Gallo and me on the prospects of discovering principles that generalize well across different biological systems and phenomena. It is often implicitly assumed that the diversity of cellular and multi-cellular behaviors results from the contingent combination of various modular parts or subprocesses, much like sequence diversity on the molecular level. This would leave us with limited avenues to pursue explanations that generalize over many such contingent assemblies.
Mulling over this in search for alternative perspectives eventually led us to an almost metaphysical argument: if there do exist principles that can explain a diverse set of biological phenomena in a unified manner, then they must be generative principles, that is to say they must comprise a mechanism by which the explained diversity is generated. But are there any such mechanisms in cellular and developmental systems, or does contingency reign supreme?
As we started looking with fresh eyes, it turned out that many of the biological phenomena we are interested in (including cytoskeletal dynamics, reaction-diffusion patterning, different aspects of multicellular morphogenesis, and even embryo-scale processes like gastrulation) can indeed be decomposed into what we have come to call a versatile core and a function-specifying periphery. A versatile core is a mechanism that implements a generative principle and hence is capable of producing a wide range of different behaviors or outputs. The periphery, then, is what configures such a core to produce one particular function out of the many in its large behavior inventory.
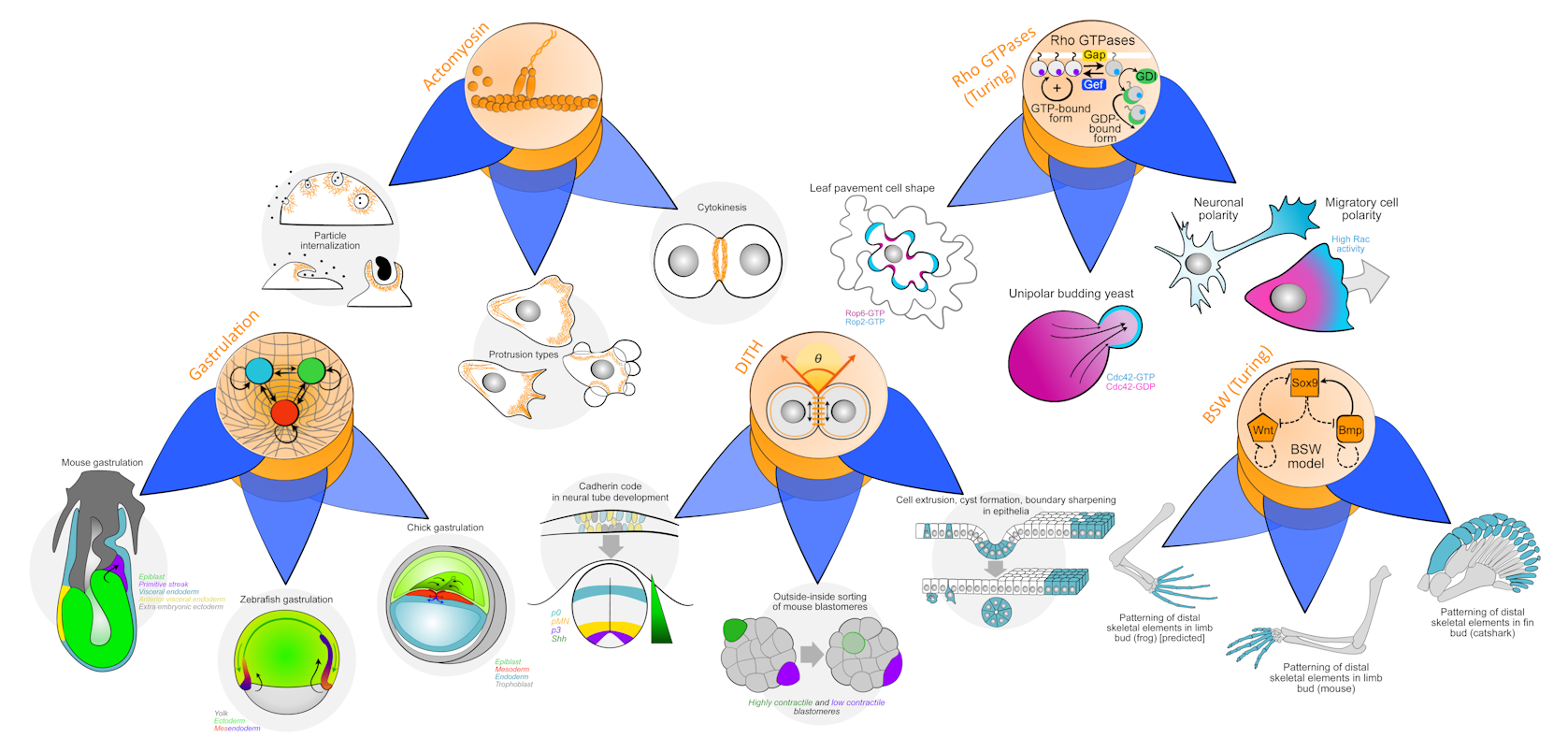
Intriguingly, we expect systems with a C&P architecture to be highly evolvable because the core’s large behavioral space is readily accessible through modifications in the periphery. As a consequence, cores will tend to spread widely and become deeply conserved in evolution, even as their peripheries diversify to exploit the full range of the core’s versatility. If follows that a generative principle that describes how a core works will generalize across the many different systems and phenomena wherein that core is reused. In other words, the C&P decomposition helps us separate the generalizable (the core) from the contingent (the periphery).
A more systematic introduction and comprehensive discussion of what the C&P hypothesis proposes is of course found in the paper. For my ramblings here, what matters most is that working on this project has greatly increased my optimism, to the point where I now believe that it really is possible to discover human-interpretable yet powerful theories that capture the essence of complex living systems. It’s just that the structure of such theories may need to differ considerably from that of the classical mechanistic accounts we are accustomed to, which is what makes it so hard (and so exciting) to pursue them.
This pursuit requires conceptual work, which means reading widely, thinking deeply, and engaging in intense and interdisciplinary discussion. As it turns out, this is surprisingly difficult and time-consuming; it is real scientific work. Unfortunately, our current research ecosystem does very little to incentivize and support such efforts. Young researchers in particular feel the pressure to pipette and/or code as fast as we can, just to stay in place in an ever-accelerating academic rat race. Taking time to think outside established lines seems wasteful, let alone taking time to pursue an explicitly conceptual project. In my case, it was only through a combination of luck, privilege, and the generosity of a few individuals that I was able to take a sabbatical year and invest the time necessary to arrive at the C&P hypothesis as it now stands. If we want the pace of conceptual innovation to pick up, this will need to change. Fortunately, there are positive signals here, too, as some leading institutes are now building up new theory-focused research programs.
In conclusion, I see many serious obstacles that we must face on our quest to better understand the complexity, intelligence, and beauty of cells and embryos. But if we take these obstacles seriously, I dare hope that we can overcome them, and that the dawn of a new golden age is indeed on the horizon.
Many thanks to Elisa Gallo and Matyas Bubna-Litic for their feedback on a draft version of this post.
[1] C.D. Stern, Reflections on the past, present and future of developmental biology, Developmental Biology 488 (2022) 30–34. https://doi.org/10.1016/j.ydbio.2022.05.001.
[2] J.B. Wallingford, We Are All Developmental Biologists, Developmental Cell 50 (2019) 132–137. https://doi.org/10.1016/j.devcel.2019.07.006.
[3] P. Liberali, A.F. Schier, The evolution of developmental biology through conceptual and technological revolutions, Cell 187 (2024) 3461–3495. https://doi.org/10.1016/j.cell.2024.05.053.
[4] D. Arendt, J.M. Musser, C.V.H. Baker, A. Bergman, C. Cepko, D.H. Erwin, M. Pavlicev, G. Schlosser, S. Widder, M.D. Laubichler, G.P. Wagner, The origin and evolution of cell types, Nat Rev Genet 17 (2016) 744–757. https://doi.org/10.1038/nrg.2016.127.
[5] A.J. Tarashansky, J.M. Musser, M. Khariton, P. Li, D. Arendt, S.R. Quake, B. Wang, Mapping single-cell atlases throughout Metazoa unravels cell type evolution, eLife 10 (2021) e66747. https://doi.org/10.7554/eLife.66747.
[6] E. Gallo, S. De Renzis, J. Sharpe, R. Mayor, J. Hartmann, Versatile system cores as a conceptual basis for generality in cell and developmental biology, Cell Systems (2024). https://doi.org/10.1016/j.cels.2024.08.001.


 (No Ratings Yet)
(No Ratings Yet)