Behind the paper: “Temporal variability and cell mechanics control robustness in mammalian embryogenesis”
Posted by Bernat Corominas-Murtra, on 15 December 2024
The massive presence of disorder and variability challenges the traditional metaphor of the developmental process as a perfectly executed program leading to precise mechanisms at every level [1,2]. Yet, the final outcome —the organism— remains both astonishingly complex and remarkably reproducible. This paradox piqued the interest of Dimitri Fabrèges and Takashi Hiiragi. Back then, around 2017, Takashi was research group leader at the EMBL Heidelberg, and Dimitri a postdoc in his group. They began to explore the idea of disorder and variability from a provoking viewpoint: instead of undermining the precision of the developmental process, randomness and variability might actually act as driving forces that ensure precision and reproducibility.
Motivated by this hypothesis, the researchers focused on the early stages of mammalian development; particularly, on the initial cleavage process of mouse, rabbit and monkey embryo, encompassing the first cell divisions post-fertilization up to the 16-cell stage. A first analysis showed that the division times of different cells in the same embryo were progressively being desynchronized. A pivotal moment in this sequence is the 8-cell stage —that is, after 3 division rounds— a moment in which cells already divided in a quite disorganized way. Due to this high variability, researchers found that the beginning of this stage was characterized by a highly heterogeneous set of cell packing configurations. However, such initial variability is smoothly but steadily reduced along the so-called compaction process, leading to a seemingly common, spherical-like structure at the end of the stage. Such a structure guarantees that, in the next round of divisions —i.e., at the 16-cell stage— there will be a suitable proportion of inner and outer cells. Achieving this correct proportion is essential: inner cells will lead to the organism itself, while outer cells form the placenta and extra-embryonic material. This observation raised a challenging question, namely, how can one support the intuitive claim that embryos begin highly heterogeneous but become remarkably similar with a more rigorous foundation.
This was the perfect challenge for Virginie Uhlmann, an expert on biological image processing who, at the time, had just started as research group leader at the EMBL-EBI, in Cambridge. She tackled this question by developing an advanced computational framework able to analyze and track the geometric changes in the embryonic shape in high detail. This approach conceptualized an embryo’s developmental path as a trajectory within a high-dimensional space whose coordinates captured relevant geometrical properties [3]. The key result was that, indeed, the trajectories exhibited significant initial disparity but converged surprisingly by the end of the compaction process in a particular region of the abstract space that characterized the embryo geometry.
Which structure was represented in this region? Why did this particular structure seem to act as the developmental “target”? At the beginning of the winter of 2018, Takashi’s research group organized a retreat in the Catalan coastal town of Sitges, gathering several groups from the ISTA. Among the attendees were Edouard Hannezo, who just opened his research group as PI, and his first postdoc, Bernat Corominas-Murtra —both physicists working on biological problems. The evening was windy and stormy, and a little café was the refuge where they largely discussed with Dimitri and Takashi about the challenge of identifying and explaining the emergent structure. Although no immediate solution came out, Edouard and Bernat concluded that a deeper and simpler structural characterization was necessary —that is, complementing the geometric analysis with a topological one, stripping out all details but the raw structure. Weeks after, they stumbled upon a relatively recent publication showing a key mathematical finding: there are exactly different 13 ways to pack 8 spheres such that none of them exhibit independent movement. This result provided the key to define a classification scheme: either the embryos conform approximately to one of these 13 packing configurations, or they are floppy, meaning that some cells retain independent movement [4]. By establishing a suitable notion of “distance” among sphere packings, the researchers could classify embryos at various developmental time points. Their analysis revealed that, although variability was very high at the onset of the 8-cell stage, as the compaction process progressed, embryos consistently converged towards these similar packing structures along similar developmental pathways. At that point, the target structure was identified: the D2d packing of 8 spheres, in the Schoenflies notation.
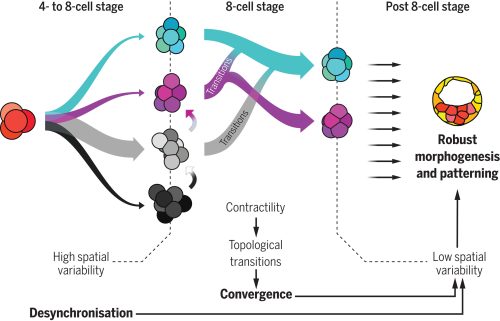
How does the embryo, without any external help, solve this kind of Rubik’s cube, i.e., transition from an arbitrary cell configuration to a specific optimal only one through successive cell rearrangements? Looking at the empirical data, one observable stood out above the other due to its clear trend: Adhesion was increasing along the compaction. Edouard suggested to challenge the simple hypothesis whether this slight change in the cell adhesion was enough to trigger all the topological rearrangements. The hypothesis has deep consequences. It implies that an increase on the cell adhesion could not only trigger deformations within the cells (i.e., increasing the contact surface, for example), but also qualitative reorganizations of the whole embryonic cell mass in a reproducible way. Computer simulations showed that such a genetically encoded slight increase in cell adhesion, coupled with significant random fluctuations in cell positions —disorder— was paradoxically facilitating the transition from any arbitrary packing of cells to the single optimal configuration. This hypothesis stands out as the simplest and, in the case of the mouse, it enabled even to reproduce in-silico the developmental trajectories of real embryos. In the case of rabbit and monkey, the role of other agents, like the zona pellucida —an external membrane that may exert a compressing force to the cell packing— could not be fully discarded.
At this point, the puzzle of the convergence towards a common, suitable embryo configuration was solved. However, the role of the temporal variability, which was experimentally observed at the starting point of the whole project and inspired it all, remained to be understood. Using several genetic perturbations, the results were surprisingly concluding that initial variability was actually required to achieve precise convergence. In particular, embryos in which cell divisions occurred more synchronously than in the wild-type ones showed a poorer convergence at the end of the compaction process, thereby hampering the further development of the embryo. The provoking hypothesis of Takashi and Dimitri on the role of stochasticity was thus proven to be fully consistent.
The researched path was not easy: Big part of the project was carried out during the COVID-19 pandemic. In turn, during the project, Dimitri and Takashi moved to Utrecht, to the Hubrecht Institute, Virginie to the University of Zurich, and Bernat to the University of Graz. Researchers from several institutions1 provided their bits of knowledge in the multiple challenges that paved the achievement of the results, and, as in living organisms, the sum of different expertises —biology, physics, mathematics and computer science— ended up in something that was much more than the sum of its parts. As in any adventurous interdisciplinary research, moments of joy and concern alternated, sometimes without pause in between… All in all, this research provides a new, constructive interpretation of the striking amount of disorder observed along developmental stages: When coupled to the changes in cell mechanics, the interplay among them can lead to significant and precise reorganization events within embryos, paving the way for a new understanding on how complex geometries and, in general, organization patterns arise in living beings. Disorder, therefore, far from being a problem the system has to deal with, may be one of the leading forces driving the precision of organism development.
Publication:
Dimitri Fabrèges et al. Temporal variability and cell mechanics control robustness in mammalian embryogenesis. Science 386, eadh1145 (2024)
References:
[1] M. Carlson, W. Reeves, M. Veeman, Stochasticity and stereotypy in the Ciona notochord. Dev. Biol. 397, 248–256 (2015).
[2] R. Dumollard, N. Minc, G. Salez, S. B. Aicha, F. Bekkouche, C. Hebras, L. Besnardeau, A. McDougall, The invariant cleavage pattern displayed by ascidian embryos depends on spindle positioning along the cell’s longest axis in the apical plane and relies on asynchronous cell divisions. eLife 6, 1–23 (2017).
[3] R. Delgado-Gonzalo, N. Chenouard, M. Unser, Spline-based deforming ellipsoids for interactive 3D bioimage segmentation. IEEE Trans. Image Process. 22, 3926–3940 (2013).
[4] N. Arkus, V. N. Manoharan, M. P. Brenner, Minimal energy clusters of hard spheres with short range attractions. Phys. Rev. Lett. 103, 118303 (2009).
1Other institutions involved:
Institute for the Advanced Study of Human Biology (WPI-ASHBi), Kyoto University, Kyoto, Japan.
Department of Developmental Biology, Graduate School of Medicine, Kyoto University, Kyoto, Japan.
Research Center for Animal Life Science, Shiga University of Medical Science, Shiga, Japan.
INRAE, BREED, Paris-Saclay University, Jouy-en-Josas, France.
École Nationale Vétérinaire d’Alfort, BREED, Maisons-Alfort, France.


 (4 votes)
(4 votes)