BSDB Gurdon Summer Studentship Report – Maciej Żurowski
Posted by Maciej Zurowski, on 27 November 2021
My glimpse into systems biology
I was introduced to the ideas of systems biology during my first year of Natural Sciences at Cambridge University. The interplay between modelling and data collection was very appealing to me. Thanks to one of my supervisors – Tim Fulton (also a PhD student in the Steventon Lab, University of Cambridge)- I was exposed to it in the context of developmental biology. He helped me get in touch with Dr Berta Verd whose interdisciplinary approach to research enticed me. We talked a lot and came up together with my project. Due to Covid we had to modify it to include modelling, but I found it more rewarding that way!
Studying bipotent posterior progenitors in Cichlids
I researched Neuromesodermal progenitor cells (NMps). They are a very interesting population of cells, persisting beyond gastrulation to generate both mesodermal and neural fates in the late embryo. This progenitor state is characterised by coexpression of two transcription factors – Brachyury (Tbxta) and Sox2 (Henrique et al. 2015). There are inter-specific differences in their proliferation dynamics – in chick and mouse embryos they proliferate, but in zebrafish they do not (Steventon and Martinez Arias 2017). This suggests they might be tuneable during the evolution of different axial elongation patterns (Sambasivan and Steventon 2021). My project was part of a larger effort in the Verd lab to see whether this might explain axial diversity observed in Lake Malawi cichlids.
I studied the NMps in Astatotilapia calliptera and Rhamphochromis chillingali – two species of cichlids from the Lake Malawi flock. They underwent a recent radial adaptation around a million years ago. Remarkably, the genetic differences between these species are very small (between 0.1-0.25% inter-specific divergence), while the morphological differences are immense (Malinsky et al. 2018). In particular, the vertebral count differs between those 2 species, making them a very suitable experimental system for studying the evolution of axial elongation.
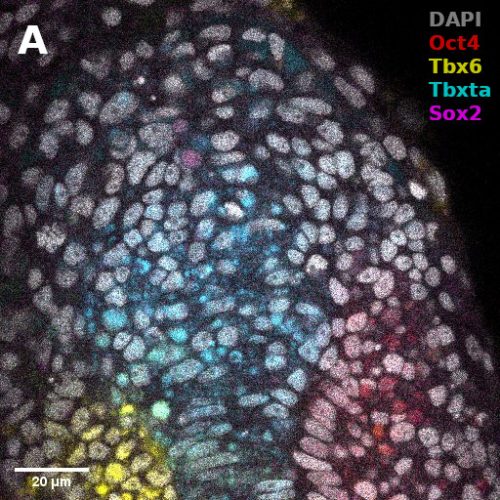
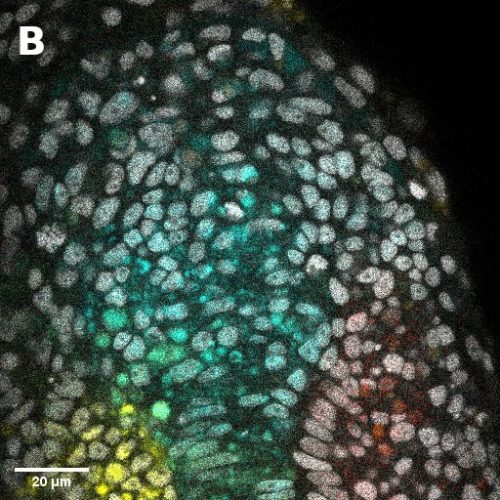
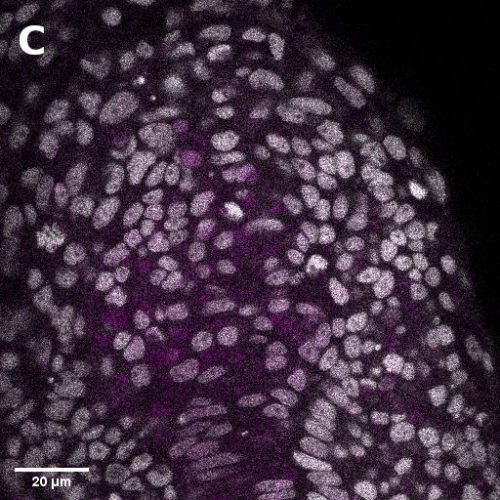
I did in situ hybridisations of the fish embryos at various stages, with the help of Shannon Taylor – a PhD student in the Verd Lab. We used Hybridisation Chain Reaction v3.0 (Choi et al. 2018). Despite our best efforts we did not manage to get Sox2 to work in the tailbud. This meant I could not quantify NMps as one of the crucial markers was missing. This was a very humbling experience – experimental biology is much more capricious than I had thought. However, it also showed me what cooperation between labs can look like. Tim did a lot of HCR staining in zebrafish at the Steventon lab in Cambridge and so we asked him for advice. It turned out it took them a few months to get Sox2 working! This was very reassuring. Tim gave us some tips, but we only managed to try some of them. Figure 1. contains some of the best images we obtained.
Modelling axial elongation
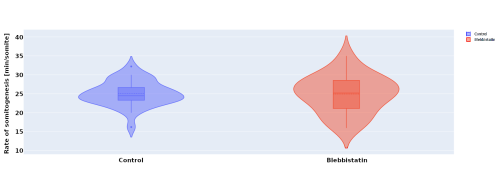
In parallel to the experimental work, I investigated the effect of blebbistatin – a myosin II inhibitor – on somitogenesis and axial elongation in zebrafish. In collaboration with the Steventon lab I analysed almost 100 timelapses of zebrafish embryos in order to determine if the rate of somitogenesis is influenced by blebbistatin. My analysis showed that it is not changed (Figure 2.). Other experiments with dye injection and cell tracking from the Steventon lab showed that the tail still elongates, but with limited cell mixing. This got us curious, how is that possible?
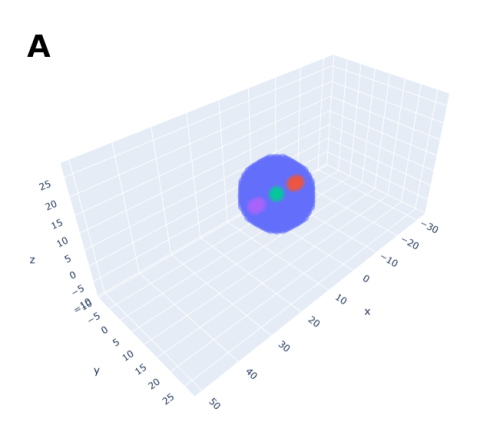
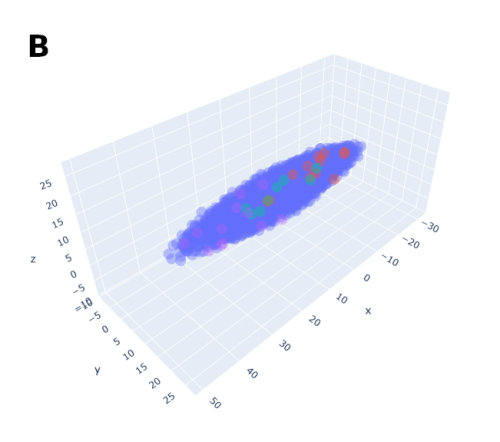
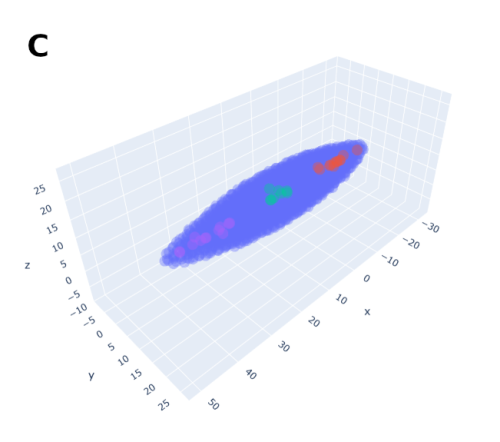
In order to help address that problem I developed a conceptual model of the zebrafish Presomitic Mesoderm (PSM) elongation. I approximated the PSM as a uniform tissue in the form of a cut-off cone. To recreate the convergence-extension mechanism responsible for axial elongation in the zebrafish tail (Thomson et al 2021; Tada and Heisenberg 2012) I gave the cells two rules for movement: they have to stay a certain range of distances apart from each other, keeping the tissues continuous and preventing cells from occupying the same space; and the cells converge towards the x-axis, mimicking the convergence movements. I found that certain combinations of parameter values indeed lead to elongation without extensive mixing, which shows that – in agreement with our experimental observations – mixing itself seems to not be required for elongation but might rather be a side effect of a certain mode of elongation (Figure 3.).
Summary
This was my first time being fully immersed in the lab. I actively took part in the lab meetings and journal clubs which were just as edifying as research itself! Overall, this was an incredible experience. It showed me that experimental biology is unpredictable and the relationship between results and time invested is non-linear. In contrast, computational biology has a much more linear relationship, almost always yielding something interesting! It also gives you the space to learn and think about underlying biological processes, how to recreate them in silico, consolidating your knowledge. This probably furthered my understanding of development the most! I am adamant I want to incorporate both experimental and computational approaches in my future research. I also gained much more understanding and appreciation of developmental biology and I want to specialise in it. I want to thank Berta, Tim, Shannon, Charlotte, Georgina, James, and Callum for welcoming me into their lab and helping me with the project, as well as BSDB for funding it.
References
Choi, Harry M.T., Maayan Schwarzkopf, Mark E. Fornace, Aneesh Acharya, Georgios Artavanis, Johannes Stegmaier, Alexandre Cunha, and Niles A. Pierce. 2018. “Third-Generation in Situ Hybridization Chain Reaction: Multiplexed, Quantitative, Sensitive, Versatile, Robust.” Development (Cambridge) 145 (12). https://doi.org/10.1242/dev.165753.
Henrique, Domingos, Elsa Abranches, Laure Verrier, and Kate G. Storey. 2015. “Neuromesodermal Progenitors and the Making of the Spinal Cord.” Development (Cambridge) 142 (17): 2864–75. https://doi.org/10.1242/dev.119768.
Malinsky, Milan, Hannes Svardal, Alexandra M. Tyers, Eric A. Miska, Martin J. Genner, George F. Turner, and Richard Durbin. 2018. “Whole-Genome Sequences of Malawi Cichlids Reveal Multiple Radiations Interconnected by Gene Flow.” Nature Ecology and Evolution 2 (12): 1940–55. https://doi.org/10.1038/s41559-018-0717-x.
Sambasivan, Ramkumar, and Benjamin Steventon. 2021. “Neuromesodermal Progenitors: A Basis for Robust Axial Patterning in Development and Evolution.” Frontiers in Cell and Developmental Biology. Frontiers Media S.A. https://doi.org/10.3389/fcell.2020.607516.
Steventon, Ben, and Alfonso Martinez Arias. 2017. “Evo-Engineering and the Cellular and Molecular Origins of the Vertebrate Spinal Cord.” Developmental Biology 432 (1): 3–13. https://doi.org/10.1016/J.YDBIO.2017.01.021.
Tada, Masazumi, and Carl-Philipp Heisenberg. 2012. “Convergent Extension: Using Collective Cell Migration and Cell Intercalation to Shape Embryos.” Development 139 (21): 3897–3904. https://doi.org/10.1242/DEV.073007.
Thomson, Lewis, Leila Muresan, and Benjamin Steventon. 2021. “The Zebrafish Presomitic Mesoderm Elongates through Compression-Extension.” BioRxiv, March, 2021.03.11.434927. https://doi.org/10.1101/2021.03.11.434927.


 (No Ratings Yet)
(No Ratings Yet)