BSDB Gurdon/The Company of Biologists 2019 Summer Studentship Report – Isabel Swinburn
Posted by BSDB, on 16 January 2020
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding. You can read accounts from previous years here. If you’re interested in applying or hosting a student in 2020, applications need to be in by the end of March.
Our third report from the class of 2019 comes from Isabel Swinburn (University of Birmingham) who studied zebrafish muscle development with Robert Knight’s lab at King’s College London.
Throughout my Biological Sciences degree, I came to realise that I want to pursue a career in medicine that combines clinical practice with research. After having thoroughly enjoyed my final year research project that focused on bacterial genetics, I decided to apply for a studentship in a biomedical research lab. This was with the hope that I’d be able to apply some of the experimental techniques I’d already learnt to understand complex processes in living organisms. Having a good grounding in organismal biology will be useful when applying a research perspective to human systems.
I was very fortunate to complete my Gurdon/The Company of Biologists Summer Studentship under the supervision of Dr. Robert Knight at King’s College London. His group uses zebrafish as a model to explore the molecular control of muscle repair and the regulation of muscle stem cells, otherwise known as satellite cells. These cells are characterised by their expression of the pax7b transcription factor (1). The zebrafish is a suitable model organism for their investigation, as it has a well-defined genome and is transparent in its larval stages. Therefore, genetic manipulations that affect cell and tissue dynamics can be readily visualised. Their work particularly appealed to me, as I am very interested in the use of reverse genetics approaches to investigate genetic diseases and how they arise due to the disruption of development.
Background to project:
The transmembrane Ret tyrosine kinase receptor is activated by the binding of Glial-derived neurotrophic factor family ligands (GFL) to Glial-derived neurotrophic factor receptors (Gfra), as this interaction stimulates Ret dimerisation and subsequently, its activation. Ret signalling has an important role in the development of the enteric nervous system and hepatic tissue, but its role in muscle development is poorly understood. Ret signalling has been implicated in facioscapulohumeral muscular dystrophy, which manifests as the weakening of the facial muscles in its initial stages (2). The binding of artemin2 (artn2), a GFL, to Grfa3 is an interaction required for the activation of Ret related to the development of the cranial muscles in zebrafish (3). My project set out to investigate the role and level of importance of Ret signalling in the development of cranial muscle satellite cells.
Experiments
1) Is an activating ligand for the Ret receptor able to alter cranial muscle development?
To test this hypothesis, I performed MF20 immunolabelling on zebrafish embryos exhibiting heat-shock inducible artn2 overexpression, as well as controls. Diaminobenzidine (DAB) staining was used to detect the signal from the antibodies, which made myofibres appear orange-brown (Figure 2a). The embryos were fixed and imaged using bright-field microscopy. The sizes of a subgroup of cranial muscles were measured and compared between the experimental and control samples (Figure 1). There did not appear to be a significant difference in muscle size between the two sample groups, suggesting that the activating ligand for the Ret receptor cannot alter cranial muscle development.
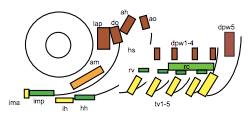
2) Is an activating ligand for the Ret receptor able to alter cranial muscle satellite cell formation?
To test this hypothesis, a transgenic line was used, in which pax7b-expressing cells were labelled with EGFP and artn2 was overexpressed. Embryos with this genotype, along with GFP+ embryos with the wild type artn2 genotype, were fixed and imaged using confocal microscopy. The number of GFP+ myofibres and myoblasts within the muscles of interest were quantified and compared between the experimental and control samples. I was unable to discriminate a significant difference, due to the control sample size being too small to be able to perform statistical tests. Therefore, more wild-type embryos need to be imaged and analysed to confirm whether artn2 can alter cranial muscle satellite cell formation.
3) How important are satellite cells for cranial muscle development?
To answer this question, I ablated pax7b-expressing cells during development. A GAL4;UAS system was used, in which the pax7b promoter induced expression of nitroreductase (NTR). Embryos which carried this system were treated with metronidazole, which reacts with NTR to produce a cytotoxic compound. The muscles of these embryos, as well as controls, underwent MF20 immunolabelling and these signals were detected using either DAB or tyramide FITC, to which a green fluorophore is conjugated. Depending on the detection method, the embryos were imaged using bright-field or confocal microscopy, respectively, and the myofibres in each image were quantified (Figure 2). Analysis of the bright-field images suggested a difference in the number of myofibres within the cranial muscles of the experimental and control sample groups. However, quantification analysis of the confocal images did not reveal a significant difference. Therefore, further experiments must be carried out to confirm the importance of satellite cells for cranial muscle development.
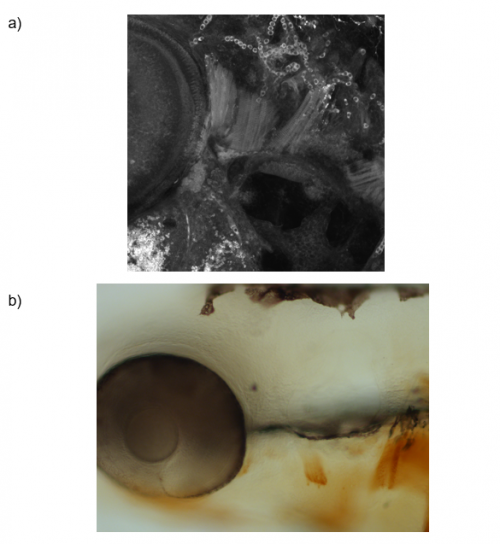
Outcomes:
At university, I had only used optical microscopy in practical sessions. However, following my time in the Knight lab, I now feel confident in using a much wider range of more advanced microscopy techniques to gather experimental data. I also appreciate much more the importance of analysing the data – quantifying cells and myofibres within images seemed like a simple task, but it took me weeks! Unlike at university, the data I collected whilst working in the lab was raw and unclean and strict parameters had to be set for its analysis. Although the task was daunting and sometimes difficult, it was very gratifying when the numbers and statistical test results allowed me to test the hypotheses generated and realise that I had made a scientific discovery.
I would like to say thank you to Robert for allowing me to work under his supervision, and to the British Society for Developmental Biology for providing the financial backing to enable me to do so. A special thank you also goes to the rest of the Knight group for all of their support and encouragement throughout. Despite my focus being primarily on a career in medicine, this experience has reinforced my desire to continue to contribute to the type of evidence that will drive medical practice. I would recommend anyone considering a career in biological research to apply for this programme.
References
- Pipalia, T. G. et al., 2016. Cellular dynamics of regeneration reveals role of two distinct Pax7b stem cell populations in larval zebrafish muscle repair. Diseases Models & Mechanisms, 9(6), pp. 671-684.
- Moyle, L. A. et al., 2016. Ret function in muscle stem cells points to tyrosine kinase inhibitor therapy for facioscapulohumeral muscular dystrophy. eLife, e11405.
- Knight, R. D. et al., 2011. Ret signalling integrates a craniofacial muscle module during development. Development, 138(10), pp. 2105-2024.
- Schilling, T. F. & Kimmel, C.B., 1997. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development, Volume 124, pp. 2945-2960.



 (No Ratings Yet)
(No Ratings Yet)