BSDB Gurdon/The Company of Biologists 2019 Summer Studentship Report – Réiltín Ní Theimhneáin
Posted by BSDB, on 21 January 2020
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding. You can read accounts from previous years here. If you’re interested in applying or hosting a student in 2020, applications need to be in by the end of March.
Our eighth report from the class of 2019 comes from Réiltín Ní Theimhneáin (NUI Galway) who studied cnidarian reprogramming in Uri Frank’s lab at NUI Galway.
Slightly different to many of the fortunate students supported by the BSDB, I do not come from an especially strong scientific background. Approaching my final year of undergraduate medicine, I wanted to spend the summer studying something not offered by the traditional medical electives. I wanted a better appreciation for the more fundamental cellular processes that underly developmental biology. I wanted some sort of foundation to work from, so I could better understand some of the science that may eventually be translated to medicine in the future. I was also curious, how would I fare out of my natural habitat of wards and surgical theatres; would I take to laboratory work, or spend my 8-weeks fumbling with protocols and pipettes?
I began my internship at Prof Uri Frank’s laboratory in the Biomedical Sciences building at NUI Galway. Under the supervision of postdoctoral researcher, Dr Miguel Salinas-Saavedra, I was given a crash course in laboratory techniques, methods to correctly follow protocols and the various rules surrounding the culture and manipulation of Hydractinia. During my internship I was expected to develop my practical skills and have an appreciation and understanding of the role of cellular senescence in the reprogramming of Hydractinia.
Hydractinia, or ‘tiny jellyfish’ as my family dubbed them, are a cnidarian model organism that has been employed in development, regeneration, and allorecognition studies. Roughly 8 mm in length Hydractinia are naturally found on the shells of hermit crabs. Prof Frank’s laboratory is the only one in Ireland to study these animals, and one of less than 10 laboratories worldwide. Similar to the freshwater cnidarian Hydra, Hydractinia have remarkable regenerative capacities. Figure 1 shows a photo of an anotated feeding polyp.

Hydractinia possess a population of adult stem cells, known as interstitial cells or i-cells. These cells are highly proliferative and normally replace cells during tissue homeostasis and regeneration1. I-cells are anatomically restricted to certain areas in the adult; they are excluded from the intact head (but can migrate to it upon injury). Interestingly, isolated heads that lack i-cells, can nevertheless regenerate a fully functional animal that includes germline competent i-cells.
The mechanisms of conversion from terminally differentiated head cells to stem cells (i.e. i-cells) is currently unknown. This idea fascinated me. The concept also represented an opportunity to study the mechanisms that can destabilize the fate of animal cells in a regenerative, non-malignant context. Preliminary work done in the Prof Frank’s laboratory suggests cellular senescence might be a potential trigger that induces dedifferentiation in neighbouring cells. This information lead to lots of reading; I needed a better understanding cellular senescence and cell cycle limitations before I started practical experiments!
At the start of my internship I tried a number of different experiments centred around cellular senescence in Hydractinia. Using the fluorescent probe SPiDER-ßGal to detect β-galactosidase activity, we sought to detect potentially senescent cells (a hallmark of senescence is high β-galactosidase activity that can be detected using SPiDER-βGal). Having detected β-galactosidase activity, Dr Salinas-Saavedra sought to optimise a method of performing an in vivo time-lapse on the confocal microscope. I meanwhile conducted some visual morphological studies in which I cut hypostomes and kept them in sea water with 5% cell culture medium, changed at regular intervals, to see the time frame associated with complete regeneration. The ultimate goal was to form fully regenerated polyps from cut hypostomes and go on to prove that these polyps were fully functional and could reproduce sexually. As the time passed we decided that the concentration of the media was too low, and the regime was changed to 10% medium for 1h changed daily. Unfortunately, by day 45 all samples had expired. Fig 2 shows an example of a healthy and an expired sample. Due to my limited time at the lab, optimising the protocol proved to be an impossible task but the experiment will be replicated by the lab, perhaps with shorter daily medium exposure.
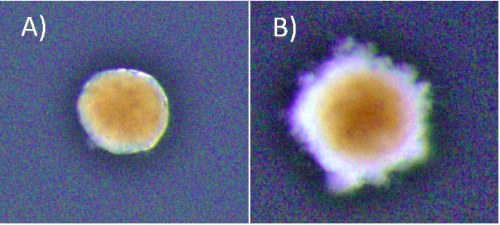
* Brightness in the photos was increased +40% for viewing purposes.
We also wanted to form a timeline to establish when cell replication occurs in hypostomes of Hydractinia at various days post cutting. To do this I conducted a double staining experiment in which I stained for both EdU and Piwi1 protein, using hypostomes at days 0-7 post-cutting along with full polyps to be used as controls. EdU is used to assay DNA synthesis in tissues and to detect cells which have undergone DNA synthesis during time of incubation. Unfortunately, EdU staining proved to be difficult in my particular case. Initial experiments failed to stain, despite positive results for the antibody and DAPI staining; the latter stains DNA. We hypothesised that I had cut the control polyps too high from the stolon (above the proliferative zone) to see any EdU staining or the EdU protocol simply had not worked. Additional issues presented with poor staining due to tissue contraction.
To try and mitigate this problem I used a new EdU and antibody staining protocol. For this I used feeding polyps cut at the stolon. To combat the contraction issue, I added MgCl2 for 15 minutes incubation prior to fixation to one sample, and sea water to the other as a control. Figure 3 shows an example of a feeding polyp from either sample group stained with DAPI, EdU and Piwi1 viewed under a confocal microscope. Interestingly, although very little change was seen in terms of tissue contraction, EdU staining appeared much stronger in the control sample in comparison to the MgCl2 exposed sample. It would have been interesting to see whether a longer time in MgCl2 would yield a more notable difference in tissue contraction.
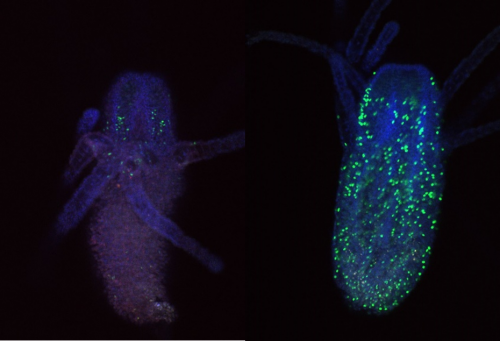
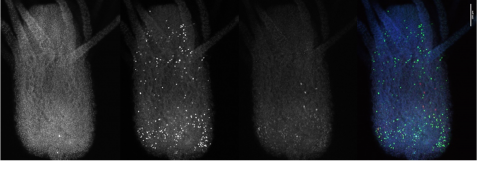
I also looked closly at the Piwi1 staining. PIWI proteins are a germline marker in hydractinia3. Staining for PIWI1 in these polyps enabled me to look at the i-cells; checking their quantity and location. It was apparent that some polyps had been cut too high as the expected i-band was not present. Figure 4 is an example of the seperated strands of DAPI, EdU, Piwi1 with the far right coloured image ultimatly compiled from each layer overlapping. Given the limited data, more experiments need to be conducted to draw any strong conclusions. Ultimately, I gained very real insight into the challenges faced daily be researchers and have a new appreciation for the resilience of those conducting experiments.
The weeks felt very short as I approached the end of my internship. I would like to thank Prof Uri Frank and all of my laboratory colleagues for their endless support and incredible patience. With the support of the BSDB Grant I was able to work as part of a cutting-edge research team, perform independent experiments, participate in weekly meetings and discuss concepts with experienced researchers. I learnt specific methods and better understood protocols. The transferable laboratory skills that I have developed will certainly help me to secure a research position after graduation, and hopefully enrich my medical career. I wish the lab all the best in their endeavours to understand the fundamental questions in regenerative biology.
References
- Plickert, Günter, et al. (2012)“Hydractinia, a Pioneering Model for Stem Cell Biology and Reprogramming Somatic Cells to Pluripotency.” The International Journal of Developmental Biology, vol. 56, no. 6-7–8, , pp. 519–534, www.ijdb.ehu.es/web/paper.php?doi=123502gp, 10.1387/ijdb.123502gp. Accessed 10 July 2019.
- Gahan, James et al. (2016). The interstitial stem cells in Hydractinia and their role in regeneration. Current opinion in genetics & development. 40. 10.1016/j.gde.2016.06.006.
- Plickert, Günter, et al. “Hydractinia, a Pioneering Model for Stem Cell Biology and Reprogramming Somatic Cells to Pluripotency.” The International Journal of Developmental Biology, vol. 56, no. 6–8, 2012, pp. 519–34, www.ncbi.nlm.nih.gov/pubmed/22689372, 10.1387/ijdb.123502gp. Accessed 10 July 2019.



 (No Ratings Yet)
(No Ratings Yet)