A day in the life of a cricket lab
Posted by Yhamada, on 14 July 2016
I am Yoshimasa Hamada, a Research Fellow in Okayama University Graduate School in Japan, working with Prof. Kenji Tomioka, Prof. Hideyo Ohuchi, Prof. Sumihare Noji and Dr. Tetsuya Bando. Our research focuses on the molecular mechanisms underlying leg regeneration, embryonic development, and circadian rhythm using the two-spotted cricket, Gryllus bimaculatus (Figure 1).
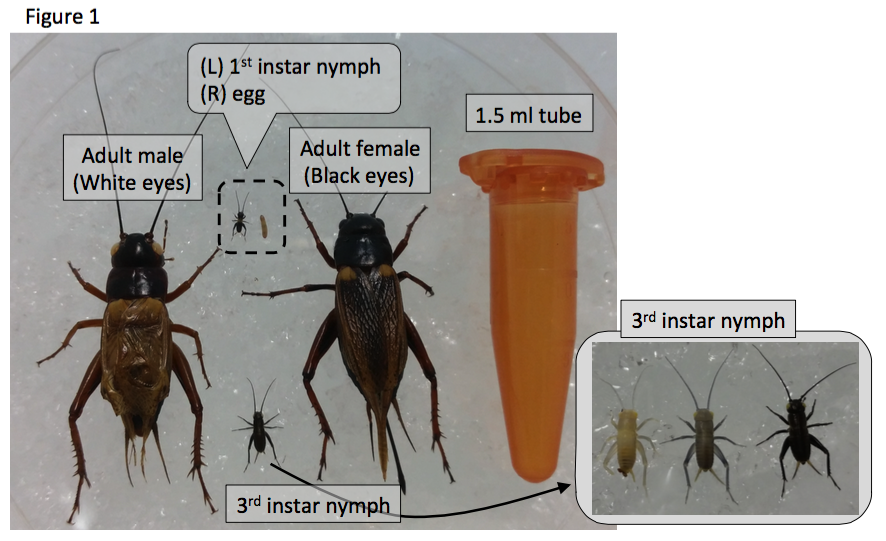
The two-spotted cricket, Gryllus bimaculatus
The two-spotted cricket Gryllus bimaculatus (De Geer) was newly described by Baron Charles de Geer in 1773. The latin name Gryllus means cricket, and bi-maculatus means two-spots, because Gryllus bimaculatus has a white spot in each forewing. The cricket has a global distribution, being found in European, African and Asian countries. Gryllus bimaculatus (and the house cricket Acheta domesticus) is a popular food source of meat-eating arthropods, amphibians, reptiles and other pet animals. Hence, we easily buy the crickets from a pet shop at a low price (we can buy 5~10 crickets for £1). Interestingly, the cricket will be a diet even for us as well as for pets: the Food and Agriculture Organization of the United Nations has proposed that insects would be a good food source for human. In fact, BBQ-flavored chips made from cricket flour are delicious for me!
The cricket is a hemimetabolous insect, with adults sized ~3cm. We easily feed them with goldfish chow and collect eggs using wet towel paper, and they hatch approximately two weeks after egg laying. Nymphs and adults are kept in the insect cages with folded paper as shelters (Figure 2). The growth rate depends on temperature: in standard breeding conditions (28 degrees Celsius), first instar nymphs hatch within two weeks, and nymphs grow up to adult by one month. All developmental stages are suitable for observation of detailed morphology, behaviors and gene expression during development.

Since crickets are relatively big and in comparison with other insects have strong vital force (which is suitable for dissection to detect action potential from targeted neurons), they have been used as a model insect for physiological research. Neuronal activities are measured by electrophysiological methods combined with pharmacological approaches.
In 1998, double-stranded RNA (dsRNA)-mediated interference (RNAi) was reported as a useful method for gene silencing by C. elegans researchers. RNAi is widely applicable for other organisms, including insects. Professor Noji’s group in Japan reported that RNAi is a powerful tool to analyze gene function during cricket embryonic development. His group also established transposon-based transgenics and genome editing techniques using zinc finger nucleases, TALE nucleases (TALEN) and CRISPR/Cas system (Nakamura et al., 2010; Watanabe et al., 2012), and analyzed whole genome sequence using next generation sequencing in Gryllus. These new methods established for the cricket open new avenues to understanding genetic basis of embryogenesis, regeneration, circadian rhythm, neurobiology, oogenesis, and so on.
Recently, we reported that epigenetic modifiers regulate leg regeneration (Development), embryonic development (Biology Open) and circadian clock (Zoological letters) by modulating specific gene expression via methylation on 27th lysine residue of histone H3. In this post, I introduce the biological features and benefits of the cricket, especially its use as a model animal in several scientific fields, e.g., regenerative and developmental biology, and chronobiology.
The cricket is a model insect of regeneration biology
Arthropods have a remarkable regenerative ability. The first scientific paper describing tissue regeneration was on the limb regeneration process of crayfish, written by René Réaumur in 1712. Hemimetabolous insects such as dragonflies, stinkbugs, grasshoppers and cockroaches were used for regeneration studies in 19th and 20th centuries. During the 1970s, the group of Prof. Vernon French documented limb regeneration and intercalary regeneration of the cockroach leg, and proposed a theory about positional values of organs; however, no molecular analyses were carried out on the cockroach. Among insect species, methods to analyze the molecular basis of organogenesis were only established in Drosophila melanogaster until the 1990s. However, the regenerative ability of Drosophila is quite limited: it can regenerate only imaginal discs but not adult legs or wings, and the body size of Drosophila is too small to be manipulated compared with crickets. Gryllus bimaculatus has remarkable regenerative ability (similarly to the cockroach) and methods to analyze molecular basis of regeneration is established, hence we start to analyze molecular basis of leg regeneration using the cricket.
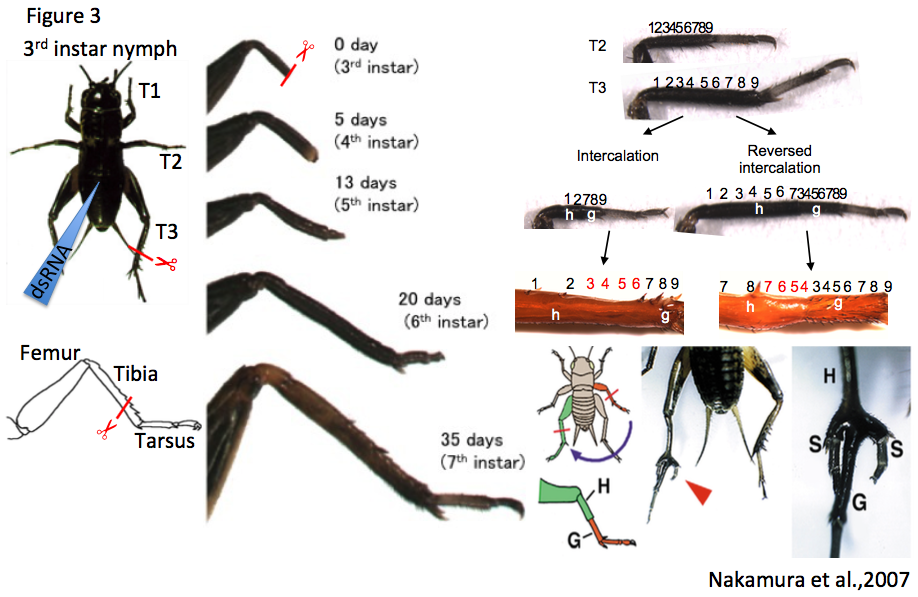
To study the molecular basis of leg regeneration, we perform RNAi in the third instar nymph and then amputate the metathoracic leg (T3) of the nymph. The crickets grow up to third instar nymphs one week after hatching at 28 degrees. Just after molting from second to third instar, the cuticle of the third instar nymphs is whitish and soft, then turns brownish and blackish, and hard by a few hours (Figure 1, window). The nymphs are around 5 mm, and look like ants. We collect whitish nymphs into 9 cm petri dish and chill on ice to anesthetize by 30 minutes. To perform RNAi in cricket nymphs, we inject dsRNA into anesthetized cricket nymphs by grass capillary held by a mechanical injector. Injecting is very easy: just insert the capillary into the abdomen of the cricket nymph and push the bottom on the mechanical injector three times (Figure 3). Then we can inject approximately 200 nL of dsRNA solution into the cricket nymph. Two days after performing RNAi, expression of endogenous target gene was decreased.
Each of the cricket prothoracic (T1), mesothoracic (T2) and T3 legs consist of the coxa, trochanter, femur, tibia, tarsus and claws, while the tarsus consists of tarsomere 1 (Ta1), tarsomere 2 (Ta2), and tarsomere 3 (Ta3). To observe regeneration process of the cricket leg, we amputate the legs using ophthalmic scissors. The lost part of the leg is completely restored by the sixth instar (within 2-3 weeks post amputation, Figure 3), and the shape of regenerated legs is indistinguishable from that of intact contralateral legs (Nakamura et al., 2008). Transplantation experiments such as intercalation, reversed intercalation, and tripod formation can be performed on cricket legs (host: T3 leg; graft: T2 leg). In the “tripod experiment”, supernumerary legs formation is induced between the host stump and graft piece (Nakamura et al., 2007) (Figure 3). To transplant a graft piece of T2 leg into the host stump of T3 leg, we just cut T2 leg at the tibia and put the T2 graft piece on the T3 host stump aligning the dorsoventral and anteroposterior axes between host and graft. Since diameters of T3 host stump is bigger than that of T2 graft piece, T2 leg easily fit to T3 leg. There is even no need to use glue to connect the host and graft. After transplantation, the cricket is put on ice 30~60 minutes to anesthetize again, avoiding dislocation of the graft piece from the host stump. The transplanted cricket will molt to next instar nymph after three days, and then newly-formed cuticle continuously covers the host stump and graft piece.
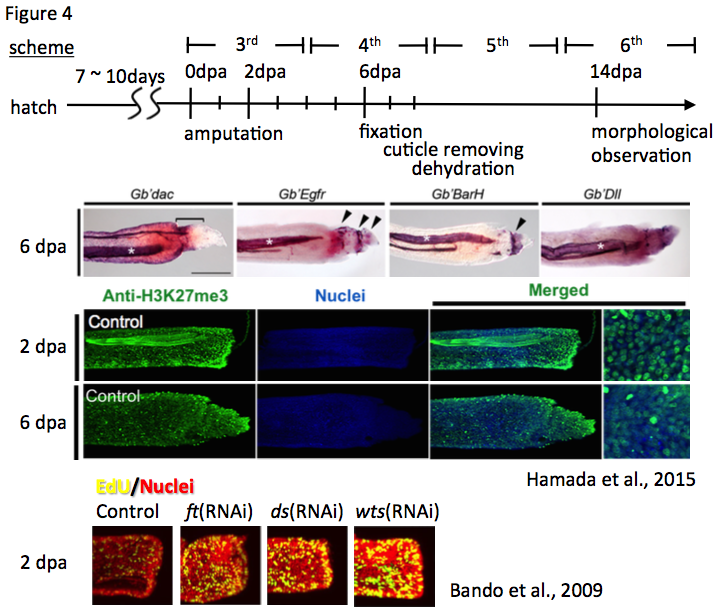
The expression pattern of target genes in the regenerating leg is detectable in the regenerating leg at two, five, or six days post amputation (dpa) by in situ hybridization. Since the cuticles of the insect inhibit penetration of RNA probes, we remove the cuticles from fixed cricket legs. The cuticles of regenerating legs at 2, 5, or 6 dpa are removed using tweezers and micro-scissors under the stereo microscope. The cuticle-removed regenerating legs can be processed for immunostaining using anti-methylated histone H3 lysine 27 (H3K27) (Hamada et al., 2015), anti-5-ethynyl-2′-deoxyuridine (EdU) (Bando et al., 2009), and anti-phosphorylated histone H3 serine 10, and so on (Figure 4).
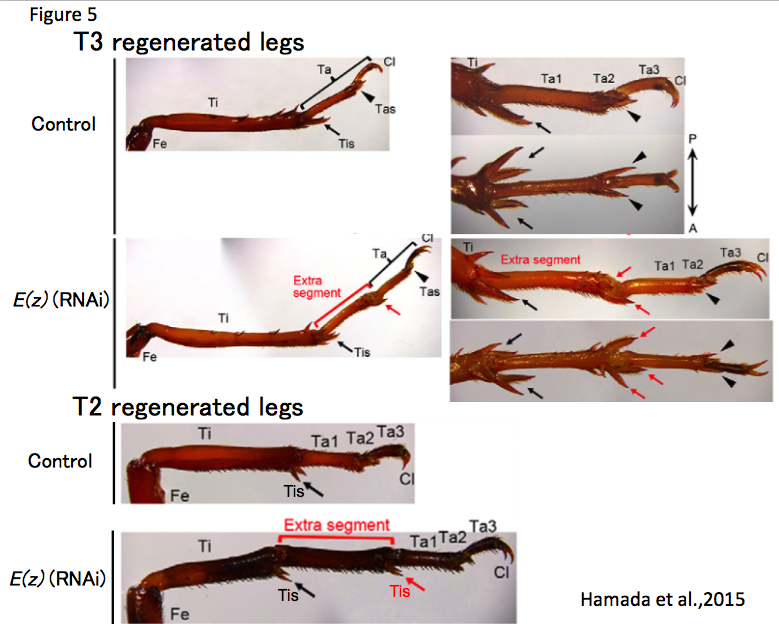
There are some morphological differences between the T2 and the T3 legs: the T3 leg has three pairs of tibial spur at the distal tibia and one pair of tarsal spur at the distal Ta1. In contrast, the T2 leg has two pairs of tibial spur at the distal tibia and has no tarsal spur at the distal Ta1. Thus, we can select the T3 leg from the T2 leg depending on the purpose of studies. For instance, when I performed RNAi against E(z) and amputation experiments at distal T3 tibia in the third instar nymph, the regenerated leg has an ectopic leg segment between the tibia and tarsus. However, I was not able to determine whether the ectopic segment is tibia or Ta1. If the ectopic segment of T2 regenerated leg has spurs at the end of the segment, we can predict that the segment has a tibia identity. In control crickets, a T2 leg was amputated at a proximal tibia in the third instar nymph, the regenerated leg restored the lost part of tissues including tibia, tibial supers, tarsus and claws. As I expected, in E(z)(RNAi) crickets, the regenerated T2 leg restored the lost part of tissue and the ectopic segment had spurs. These results suggest that the ectopic segment has a tibia identity and morphological character of T2 leg is suitable for regeneration research (Hamada et al., 2015) (Figure 5).
The cricket shows short germ segmentation
A large number of the cricket eggs are collectable at all times of the year. We supply water to the crickets using wet towel paper put into petri dishes. Adult female crickets lay eggs into the wet towel paper. An adult female cricket lays ~3,000 eggs in a few weeks. Usually we keep ~100 adult males and females, hence we can get ~100 eggs within one hour. If we quickly move the towel paper in the water, the laid eggs are removed from the paper, then we can collect the cricket eggs from the water using a tea strainer. We can inject dsRNA for embryonic RNAi, transposons for transgenesis, RNA encoding TALEN or CRISPR/Cas for genome editing, pharmacological reagents and others into the ventroposterior side of the fertilized eggs (Figure 6).
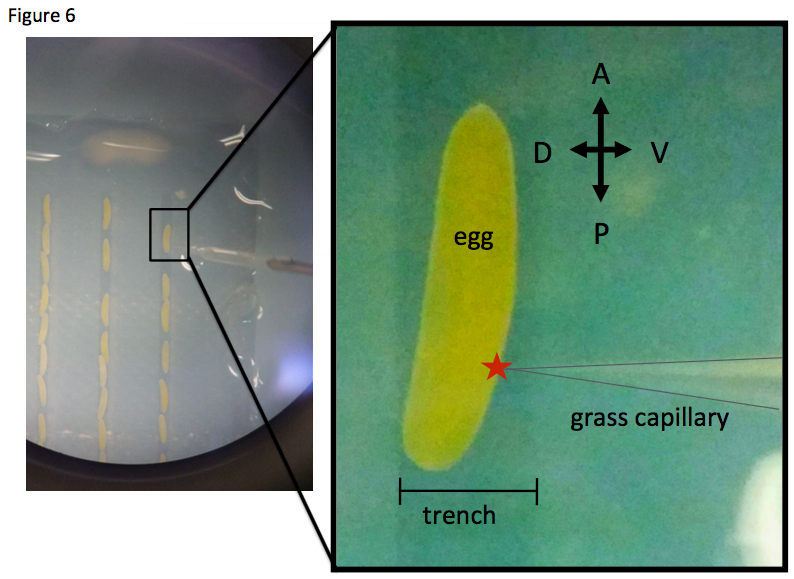
The egg diameter is approximately 3 mm x 0.5 mm. The developmental stage of cricket embryos is well documented from fertilization to hatching. The early embryo is formed on the ventroposterior side of the egg. We dissect fertilized eggs to get cricket embryos in the desired developmental stages (Donoughe and Extavour, 2016). These embryos are applicable for whole mount in situ hybridization to detect gene expression. While the regeneration processes of the leg can be observed during molting, the embryo can be continuously observed and collected anytime for molecular analysis. For efficient RNAi during early embryonic development, we inject dsRNA into the body fluid of adult female (Matsuoka et al., 2015). Early embryos in the ovary intake dsRNA, called parental RNAi. The model animal Drosophila melanogaster shows the long germ segmentation in its developmental mode, while the cricket shows the short germ segmentation in its developmental mode. Thus, in Evo-Devo studies, the cricket is expected to be a more representative model insect.
The cricket shows characteristic circadian rhythm
The cricket shows diurnal activity in nymphal stages, but after imaginal molting, the activity phase is shifted to be nocturnal (Tomioka and Chiba, 1982). Activities of cricket individuals are documented by using small cricket cages with seesaw-like flower, that is a magnetic sensor switch on the flower (Figure 7).
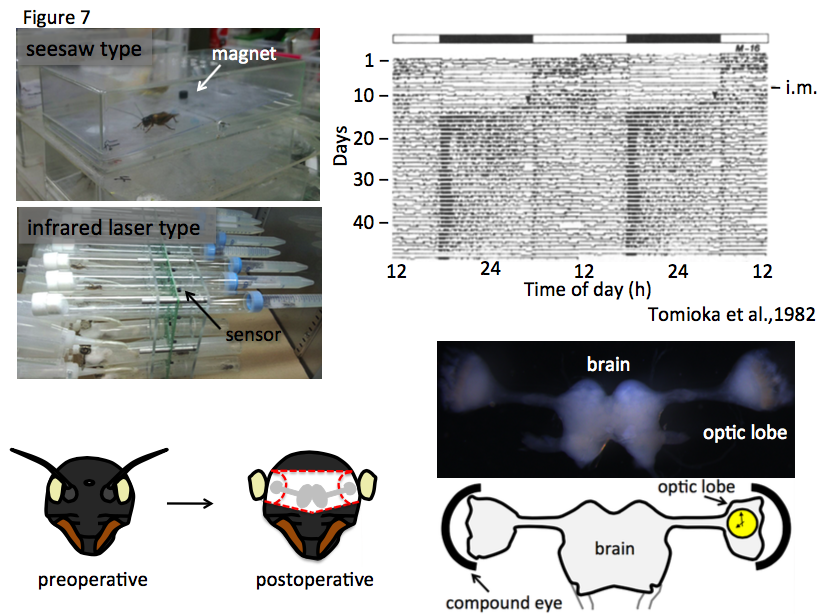
To record the actgram of more smaller animals (the body length of G. bimaculatus is less than 25mm), I have succeeded to utilize LAM25 with an infrared laser sensor for Modicogryllus siamensis (Trikinetics inc). The data from the seesaw and infrared laser locomotor recorder can be analyzed by Actogram J, a plug-in in Image J (unpublished). Control and/or RNAi-treated crickets are reared in the small cage to measure continuous actogram for more than a month. Previously, it was reported that the central clock system is located in the optic lobe (Tomioka and Chiba, 1992), where clock component gene expression is oscillated daily (Tomioka, 2014). In the cricket, the optic lobes locate bilaterally in the brain, at the inside of compound eyes. The clock genes and their expression profiles are similar to those of model animals including mice and flies. For instance, the clock component genes of mice, such as Clock, Bmal1, period1/2/3, Timeless and Cryptochrome are conserved in fly and cricket as Clock (Clk), cycle (cyc), period (per), timeless (tim) and cryptochrome2 (cry2), respectively, and the transcriptional/translational molecular feedback loops for period of 24 hours are consistent between mice and insects. When we measure the activities of per(RNAi) crickets, they show arrhythmic actogram. Furthermore, it is easy to obtain the whole brain, which is also a central clock component, and used for in situ hybridization (Hamada et al., 2009), immunostaining, and so on. Recently, we address the big challenges that elucidate the relationship between leg regeneration and photoperiodism, and systematize knock-in cricket lines using CRISPR/Cas9 system.
The cricket is one of the most familiar insects to enjoy insect songs in autumn in Japan. A Japanese author, Sei Shonagon mentioned that “Evening is the best time in the day in autumn, because we can enjoy the flight of wild geese and songs of insects” in her essay named “Makurano soushi (The Pillow Book)”, written in the 10th century. Now the cricket is used as a model insect of regeneration biology and RNAi-based analyses show new insights into tissue regeneration in addition to his song. I am thinking about new experiments listening to crickets’ chirp songs today.
References
Bando, T., Mito, T., Maeda, Y., Nakamura, T., Ito, F., Watanabe, T., Ohuchi, H. and Noji, S. (2009). Regulation of leg size and shape by the Dachsous/Fat signalling pathway during regeneration. Development 136, 2235-2245.
Donoughe, S. and Extavour, C. G. (2015). Embryonic development of the cricket Gryllus bimaculatus. Dev Biol 411, 140-156.
Hamada, A., Miyawaki, K., Honda-sumi, E., Tomioka, K., Mito, T., Ohuchi, H. and Noji, S. (2009). Loss-of-function analyses of the fragile X-related and dopamine receptor genes by RNA interference in the cricket Gryllus bimaculatus. Dev Dyn 238, 2025-2033.
Hamada, Y., Bando, T., Nakamura, T., Ishimaru, Y., Mito, T., Noji, S., Tomioka, K. and Ohuchi, H. (2015). Leg regeneration is epigenetically regulated by histone H3K27 methylation in the cricket Gryllus bimaculatus. Development 142, 2916-2927.
Matsuoka, Y., Bando, T., Watanabe, T., Ishimaru, Y., Noji, S., Popadic, A. and Mito, T. (2015). Short germ insects utilize both the ancestral and derived mode of Polycomb group-mediated epigenetic silencing of Hox genes. Biol Open 4, 702-709.
Nakamura, T., Mito, T., Bando, T., Ohuchi, H. and Noji, S. (2008). Dissecting insect leg regeneration through RNA interference. Cell Mol Life Sci 65, 64-72.
Nakamura, T., Mito, T., Tanaka, Y., Bando, T., Ohuchi, H. and Noji, S. (2007). Involvement of canonical Wnt/Wingless signaling in the determination of the positional values within the leg segment of the cricket Gryllus bimaculatus. Dev Growth Differ 49, 79-88.
Nakamura, T., Yoshizaki, M., Ogawa, S., Okamoto, H., Shinmyo, Y., Bando, T., Ohuchi, H., Noji, S. and Mito, T. (2010). Imaging of transgenic cricket embryos reveals cell movements consistent with a syncytial patterning mechanism. Curr Biol 20, 1641-1647.
Tomioka, K. (2014). Chronobiology of crickets: a review. Zoolog Sci 31, 624-632.
Tomioka, K. and Chiba, Y. (1982). Post-embryonic development of circadian rhythm in the cricket, Gryllus bimaculatus: A rhythm reversal. J Comp Physiol 147, 299-304.
Tomioka, K. and Chiba, Y. (1992). Characterization of an optic lobe circadian pacemaker by in situ and in vitro recording of neural activity in the cricket, Gryllus bimaculatus. J Comp Physiol A 171, 1-7.
Watanabe, T., Ochiai, H., Sakuma, T., Horch, H. W., Hamaguchi, N., Nakamura, T., Bando, T., Ohuchi, H., Yamamoto, T., Noji, S., et al. (2012). Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun 3, 1017.
This post is part of a series on a  day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.


 (4 votes)
(4 votes)
” …His group also established transposon-based transgenics and genome editing techniques using zinc finger nucleases, TALE nucleases (TALEN) and CRISPR/Cas system (Nakamura et al., 2010; Watanabe et al., 2012), and analyzed whole genome sequence using next generation sequencing in Gryllus….”
Is it possible to get access to this Gryllus genome sequences?