In Development this week (Vol. 143, Issue 8)
Posted by Seema Grewal, on 19 April 2016
Here are the highlights from the current issue of Development:
The origins of lung macrophages revealed

Tissue-resident macrophages are phagocytic cells that are essential for the response to injury and infection. Both within and between tissues, macrophages can show distinct characteristics, but are these attributes developmentally defined or determined by the microenvironment? In the mouse lung, there are two distinct macrophage populations: alveolar macrophages that reside within the lumen of the alveoli and interstitial macrophages that occupy the interalveolar space and elsewhere in the lung parenchyma. Some studies have suggested that alveolar macrophages originate from and are repopulated by an interstitial macrophage precursor, while others indicated that they can maintain themselves independently. Serena Tan and Mark Krasnow (p. 1318) now show that there are in fact three developmentally distinct lineages that populate the lung in three waves, with minimal interconversion between them – at least under homeostatic conditions. The first population, derived from yolk sac haematopoietic cells, populate the interstitial space in embryogenesis, but become confined to peripheral and perivascular regions postnatally. The second, of as-yet-unknown origin, initially occupy the interstitium but then become alveolar macrophages. The third population enters the lung postnatally from circulating monocytes and constitute the majority of mature adult interstitial macrophages. In the lung at least, it appears that developmental origin, rather than environmental influence, is the primary determinant of macrophage identity and diversity.
Transcription without TBP

The canonical mechanism of transcription initiation in all metazoans involves recruitment of TATA-binding proteins (TBP, TLF or TBP2 in vertebrates) to the promoter, as a rate-limiting step before binding of RNA polymerase II. TBP, TLF and TBP2 have non-redundant functions, but the degree of redundancy between them is not clear; nor is it well understood whether there are TBP-independent mechanisms of transcription initiation in vivo. On p. 1340, Gert Veenstra and co-workers now show that, during early Xenopus development, there is a small group of genes whose transcription is independent of all TBP family members (denoted TBP family-insensitive or TFI genes). These genes are enriched for factors expressed in mesoderm and at Spemann’s organiser and include several key transcription factors involved in mesendoderm specification. Strikingly, most TFI genes are bound by these TFI transcription factors. Gcn5, a component of the SAGA complex thought be involved in non-canonical transcription initiation, is recruited to TFI promoters upon TBP family knockdown, is not required for their transcription in the presence of TBP-related factors, but seems to compensate in their absence. This work provides clear evidence that alternative mechanisms of transcription initiation exist in vivo, and that they may be preferentially used for a particular set of key developmental genes.
Completing the neuroblast map

In the developing Drosophila embryonic central nervous system (CNS), the pattern of neural stem cells – neuroblasts (NBs) – is highly stereotyped, both between individuals and, in the truncal ventral nerve cord, between segments. Over the past decades, multiple studies have mapped the spatio-temporal origin and gene expression signature of the embryonic NBs in the brain, thorax and abdomen. On p.1290, Rolf Urbach and colleagues provide the final piece to this puzzle by providing a comprehensive map of the NBs in the gnathal (labial, maxillary and mandibular) segments of the embryo. In doing so, they are able to compare the NBs complement of each segment to identify homologies between NBs in different embryonic origins. Their work demonstrates the progressive loss of NBs from trunk to progressively anterior gnathal segments and analyses its cause. Despite the reduced NB number, homologies in developmental origin and expression pattern are clearly recognisable, and can also be traced into posterior brain segments. The wealth of data in this and related papers provide an essential foundation to understand the molecular and evolutionary basis of segmental diversification of the CNS.
Vascular development in technicolour

During development, the vascular endothelium becomes covered by mural cells (MCs) – vascular smooth muscle cells or pericytes – that are essential for vascular stability and homeostasis. MCs are known to be of mesodermal or neural crest origin, but little is known about how they are recruited to and cover the vessels – primarily because live imaging of this process has been challenging. Now (p.1328), Shigetomo Fukuhara, Naoki Mochizuki and colleagues overcome this hurdle by developing transgenic zebrafish lines to mark MCs fluorescently. They then use these tools to follow the origin and subsequent behaviour of MCs in both cranial and trunk regions of the embryo. The authors find that trunk MCs are mesodermal in origin, while both neural crest and mesoderm populations contribute to cranial MCs. MCs appear to be recruited to specific vessels, such as the dorsal aorta in the trunk or the basilar artery in the head, and then migrate using inter-endothelial cell junctions as a scaffold to cover other vessels – preferentially the arteries. As well as providing important insights into MC behaviour, the tools developed here should serve as a valuable resource for the community for future analyses of vascular development.
Determining dendritic diversity in Drosophila

Neuronal morphology is highly variable, particularly in terms of the complexity of dendritic arborisation, and this variability is crucial for appropriate function. But how is such diversity established and regulated? Wesley Grueber and colleagues (p. 1351) set out to address the transcriptional inputs into this process using a subset of Drosophila sensory neurons, the multidendritic (md) neurons, whose morphology is regulated by the transcription factor Cut. Cut expression is absent in neurons that have simple morphology and function as proprioceptors, but is expressed at variable levels in nociceptive or touch-sensitive neurons with more complex dendrites. Through a series of mosaic genetic analyses, the authors find that Cut represses the expression of the Pdm1/2 transcription factors in a subset of md neurons, which suppresses the ability of Pdm1/2 to restrain dendritic arborisation. Upstream of Cut, the transcriptional repressors Vestigial and Scalloped modulate Cut levels to limit dendritic elaboration – in this case repressing a complex morphology and favouring a less complex type of branching. Together, these data identify a network of repressive interactions that regulate neuronal morphology and thus help to define neuronal identity and diversity.
PLUS…
CncRNAs: RNAs with both coding and non-coding roles in development
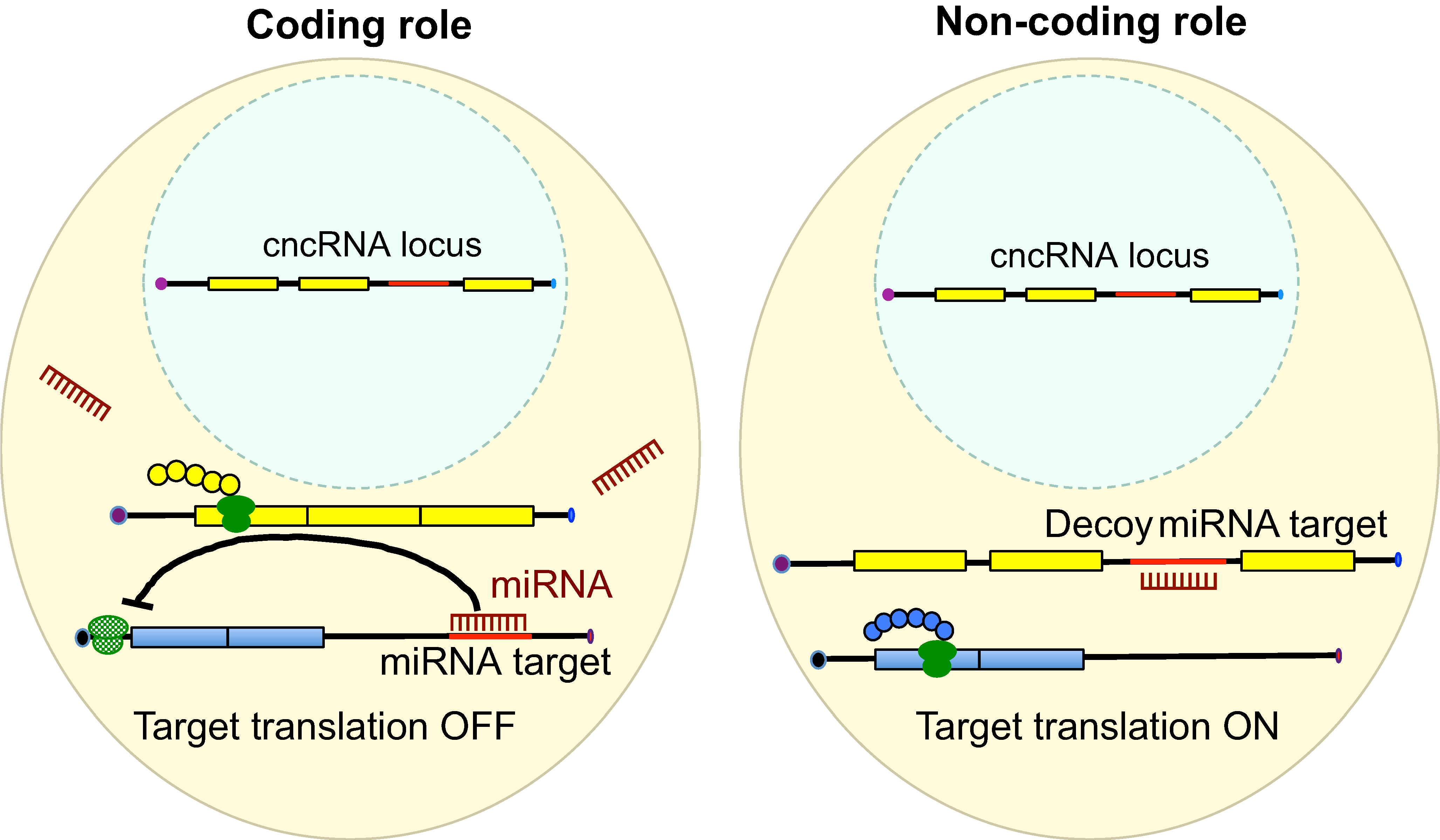 RNAs are known to regulate diverse biological processes, either as protein-encoding molecules or as non-coding RNAs. However, a third class that comprises RNAs endowed with both protein coding and non-coding functions has recently emerged. Such bi-functional ‘coding and non-coding RNAs’ (cncRNAs) have been shown to play important roles in distinct developmental processes in plants and animals. Here, discuss key examples of cncRNAs and review their roles, regulation and mechanisms of action during development. See the Primer on p. 1234
RNAs are known to regulate diverse biological processes, either as protein-encoding molecules or as non-coding RNAs. However, a third class that comprises RNAs endowed with both protein coding and non-coding functions has recently emerged. Such bi-functional ‘coding and non-coding RNAs’ (cncRNAs) have been shown to play important roles in distinct developmental processes in plants and animals. Here, discuss key examples of cncRNAs and review their roles, regulation and mechanisms of action during development. See the Primer on p. 1234
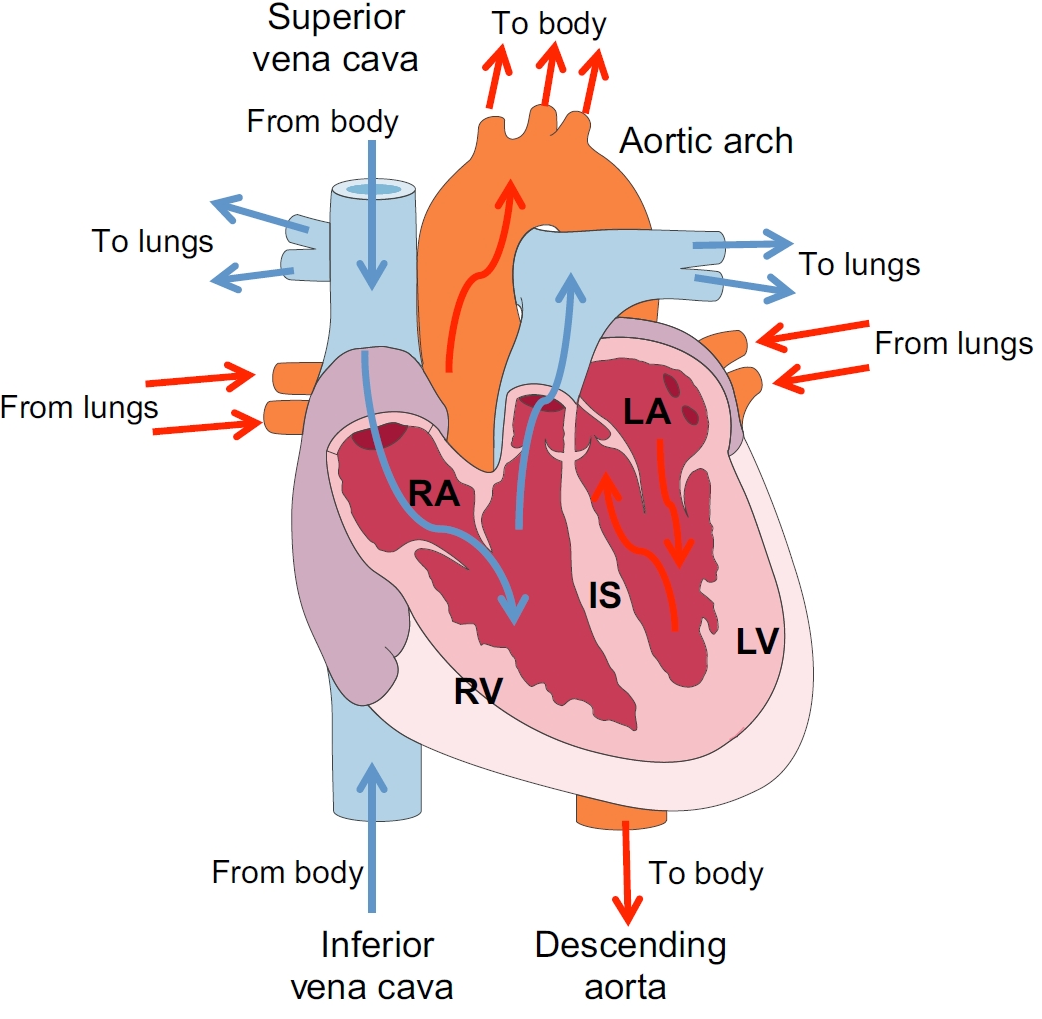 Developmental origin and lineage plasticity of endogenous cardiac stem cells
Developmental origin and lineage plasticity of endogenous cardiac stem cells
Over the past two decades, several populations of cardiac stem cells have been described in the adult mammalian heart. Here, ,
Lineage-specific stem cells, signals and asymmetries during stomatal development
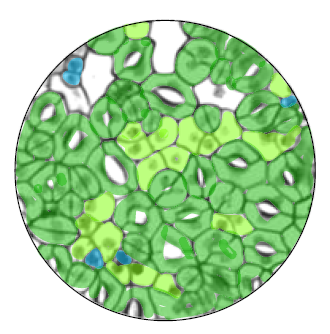 Stomata – the dispersed pores found in the epidermis of land plants that facilitate gas exchange – are formed from progenitor cells that execute a series of differentiation events and stereotypical cell divisions. Here,
Stomata – the dispersed pores found in the epidermis of land plants that facilitate gas exchange – are formed from progenitor cells that execute a series of differentiation events and stereotypical cell divisions. Here,


 (2 votes)
(2 votes)