Featured Resource: Echinobase
Posted by Cheryl Telmer, on 6 December 2022
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. Our ‘Featured resource’ series aims to shine a light on these unsung heroes of the science world. In our latest article, we hear from Cheryl Telmer, who describes the work of Echinobase.

Overview
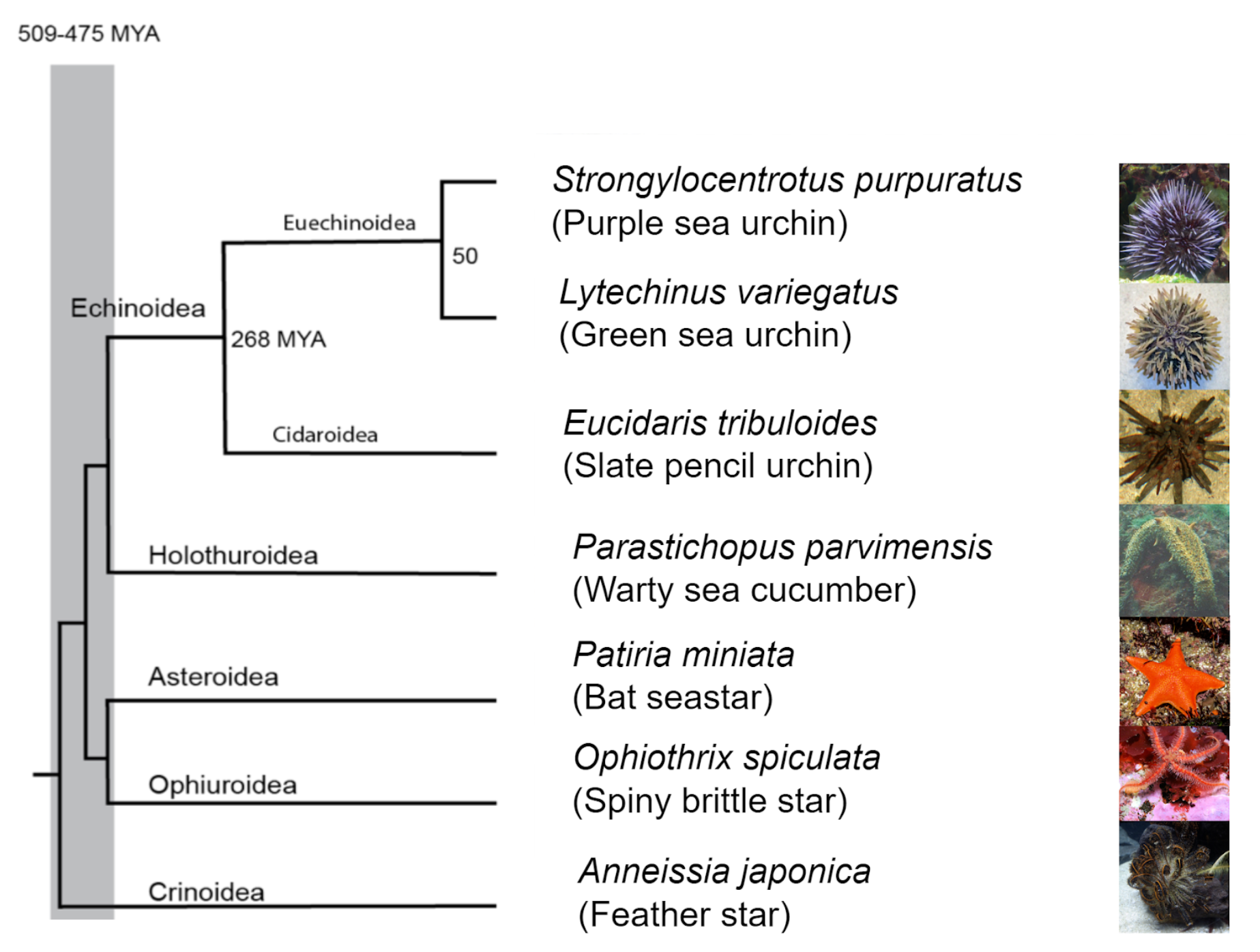
Echinobase (www.echinobase.org) is a model organism knowledgebase for the echinoderm research community (Arshinoff et al. 2022). The Phylum Echinodermata has 5 Classes: the basal branching Crinoidea (crinoids), and the 4 motile Eleutherozoa Classes including Asterozoa, composed of the Ophiuroidea (brittle stars) and Asteroidea (starfish) and the Echinozoa, composed of the Echinoidea (sea urchins and sand dollars) and Holothuroidea (sea cucumbers). Echinoderms, along with the phyla Chordata and Hemichordata are Deuterostomes. The species used in research labs have many advantages for studying embryo development and regeneration including an abundance of gametes, synchronized fertilization, transparent embryos and larvae, ease of microinjection and micromanipulation, and variation in development within and between a species. This has made echinoderms a premier model system to study gene regulation, genomic control of cell specification and gene regulatory networks in development and evolution.
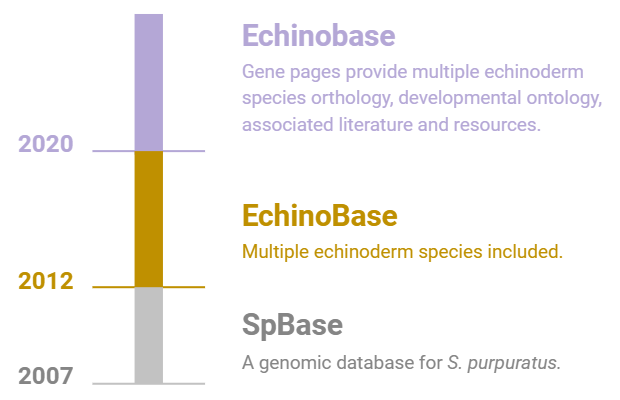
Echinobase is the third generation computational resource for the support of all genomic data and research using echinoderm model organisms.
The Echinobase Teams
Echinobase has been developed from previous databases including SpBase (previously funded by NIH R01 to PI Andy Cameron, 2007-2012) and EchinoBase (NIH P41 to PI Cameron, 2012-2018). Echinobase development is now funded through a collaborative NIH P41 between Carnegie Mellon University (PIs Hinman/Ettensohn) and the University of Calgary (PI Vize). The database and software development team are in Calgary and also run the Xenopus knowledgebase, Xenbase. The data curation and bioinformatics team are at Carnegie Mellon University, where the PIs are leading researchers in the echinoderm research community. The current Echinobase is a clone of Xenbase which has adapted multiple new features and content in support of the echinoderm community, and broader needs of the developmental biology communities.

Support for Genomics Research
The most current genome assemblies are hosted on Echinobase. Currently four species are fully supported, Strongylocentrotus purpuratus (purple sea urchin), Lytechinus variegatus (variegated sea urchin), Patiria miniata (bat star) and Acanthaster planci (Crown of Thorns sea star). Partial support (BLAST and JBrowse) is provided for Asterias rubens (sugar star) and Anneissia japonica (feather star, a crinoid). Search and BLAST tools are available on the landing page or through the over 38,000 gene pages. Gene pages also display gene model HGNC compliant names, multispecies orthology, GO terms, a link to the JBrowse genome browser, and a gene expression plotting tool. Tabs beyond the summary on the gene page provide gene specific literature, transcripts, expression data, protein sequences, interactants and more.
The Echinoderm Anatomical Ontology
The Echinoderm Anatomical Ontology (ECAO) describes echinoderm anatomy and embryological development using a controlled vocabulary of anatomy terms and developmental stages that are organized in a hierarchy with a graphical structure. Echinobase curators use ECAO terms to describe the spatiotemporal expression patterns of endogenous gene products, the transcriptional activity of cis-regulatory modules, the effects of morpholinos, and other types of biological information. The ECAO is constantly being updated in response to the latest published echinoderm research.
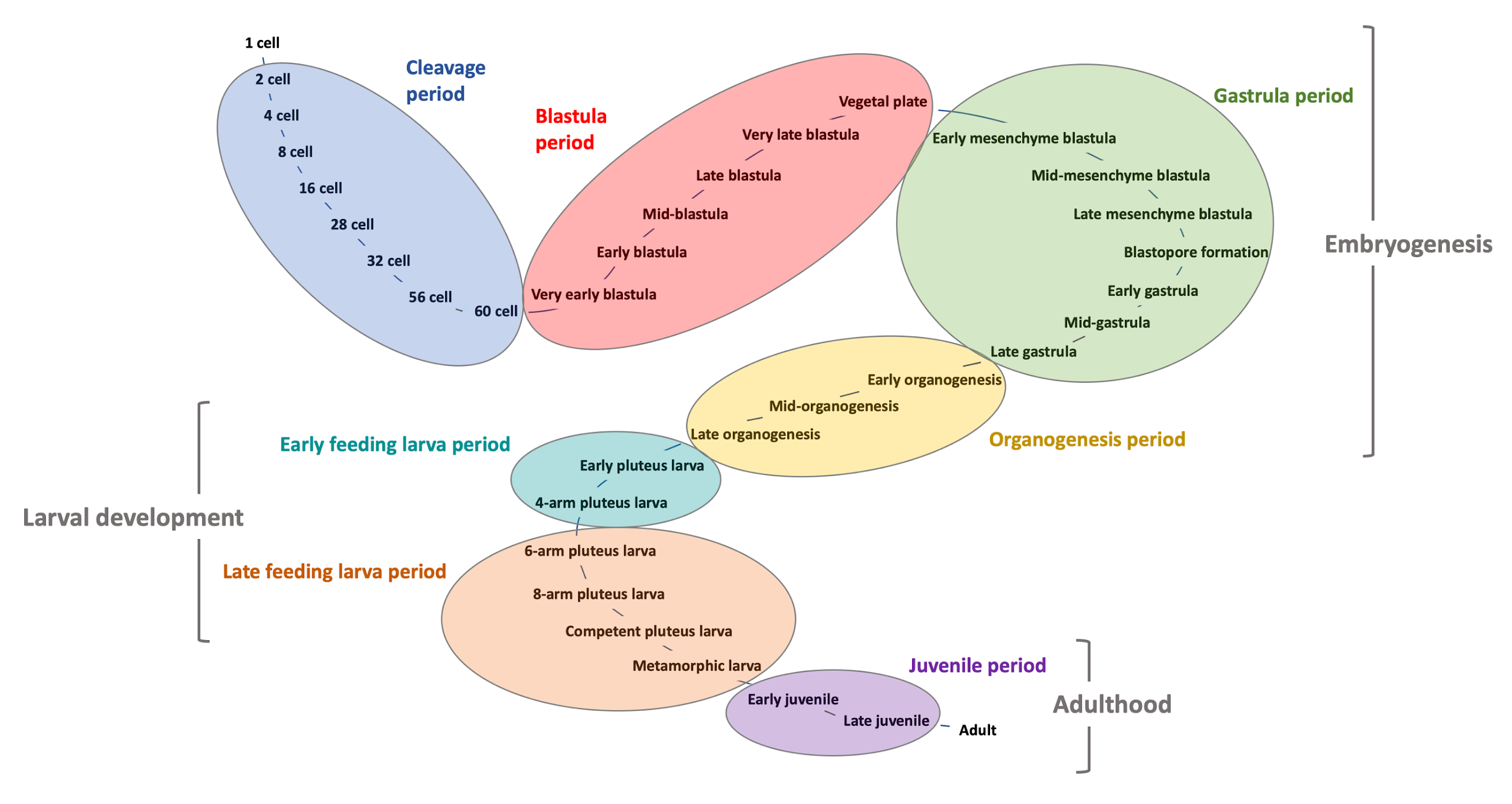
The ECAO contains 1000’s of terms and relationships. Each anatomical system (e.g. nervous system, skeletal system), tissue and structure (e.g. ciliary band, mesoderm, blastopore) and many cell types (e.g. pigment cell, serotonergic neuron, skeletogenic mesenchyme cell) are separate terms; each term is fully defined and related to other terms by is_a, part_of, develops_from and develops_into relationships. The ontology makes text and data computer readable and allows our code to link and infer relationships between many different types of content.
Manual and Automated Curation
Automated literature collection has retrieved over 18,000 publications for automated and manual curation. Text matching automatically links gene symbols or names appearing in manuscript text to gene pages. Manual curation extracts sequence information for morpholinos, gRNAs, and cis-regulatory elements and itemizes antibodies used in experiments.
Sharing on EchinoWiki and FTP
To support the community, collections of data, protocols and other resources are shared using EchinoWiki and an FTP site. To enable interdisciplinary and collaborative studies, contact information for community members and groups and information about their research are available and searchable.
Help from users
The echinoderm research community has always been extremely supportive and can continue to do so at many levels. It is highly important and appreciated if users cite Echinobase whenever possible in articles, presentations and funding applications (citation link in the top right corner of the webpage “Citing Echinobase”). These acknowledgements make Echinobase’s impact on research more tangible and specifically the article citations provide metrics that can be used for funding applications.
Help from authors
Authors can also contribute in several ways to simplify the curation of their articles, ultimately allowing their data to be more quickly available on the website.
When you write your paper…
In addition to “Citing Echinobase”, clear, detailed and accurate descriptions of the experiments and resources minimizes the curation effort and reduces the need to contact the authors. Articles should mention official Echinobase identifiers and nomenclature for entities such as genes, alleles, and anatomical structures.
Bibliography
Arshinoff BI, Cary GA, Karimi K, Foley S, Agalakov S, Delgado F, Lotay VS, Ku CJ, Pells TJ, Beatman TR, Kim E, Cameron RA, Vize PD, Telmer CA, Croce J, Ettensohn CA, Hinman VF, Echinobase: leveraging an extant model organism database to build a knowledgebase supporting research on the genomics and biology of echinoderms, Nucleic Acids Research, Volume 50, Issue D1, 10.1093/nar/gkab1005


 (1 votes)
(1 votes)