Featured Resource: Gene Expression Database (GXD)
Posted by the Node, on 5 December 2024
Our ‘Featured resource’ series aims to shine a light on the resources that support our research – the unsung heroes of the science world. In this post, Connie Smith and Martin Ringwald introduce the data and functionalities available at Gene Expression Database and talk about the future directions of the database.
What is the Gene Expression Database (GXD)?
GXD is a long-standing, freely available community resource that collects and integrates mouse gene expression data generated by biomedical researchers worldwide. Our primary emphasis is on endogenous gene expression during development, covering data from wild-type and mutant mice. Data types include RNA in situ hybridization, immunohistochemistry, knock-in reporter, RT-PCR, Western blot, and RNA-seq. Close to half a million expression images (mainly from in situ data) are readily available and accessible to searches. As an integral component of Mouse Genome Informatics (MGI), GXD combines its expression information with genetic, functional, phenotypic, and disease-oriented data, thus facilitating the study of the molecular mechanisms of development, health, and disease.
What inspired the development of GXD?
We began developing GXD in 1993 for two main reasons: (i) working in the field of mouse development, we realized that there was no database to store and integrate all the mouse expression data being published, and (ii) it was clear that gene expression information would provide a crucial link between the mouse genetic and phenotypic information already being collected by the emerging Mouse Genome Database (MGD). Our plan always was to combine MGD and GXD into the integrated resource which is now known as MGI.
The development of GXD was a pioneering effort because no comparable resources existed at the time. Our biological requirement analysis resulted in a publication in Science in 1994 and has served as a blueprint for our work ever since. The basic concepts developed there are still relevant today. This includes the discussion of the complementary nature of text-based (i.e. anatomy ontology-based) and spatial representation of expression patterns, and the notion that both approaches should be combined to integrate the data and to enable effective searching and reasoning over them.
What is available at GXD?
Since the beginning, GXD has collected classical types of mouse developmental expression data, i.e. in situ and blot data. These data are acquired through systematic curation of the scientific literature and by collaborations with large-scale expression projects.
Comprehensive literature survey. As a first step in our literature annotation work, our curators review new publications to find studies of endogenous gene expression during mouse development that use classical types of expression assays. They then index all genes that have been studied in the paper, the assay types used, and the ages analyzed. These data, combined with bibliographic information from PubMed, can be accessed using the Gene Expression Literature Search. These searches are more effective and complete than PubMed searches because the annotations are based on the entire article, including supplemental data. This index is up to date and currently includes over 33,000 publications.
Detailed expression data that are easily searchable in many ways. GXD expression annotations are detailed, making extensive use of controlled vocabularies and ontologies, as illustrated. These standardized metadata enable the close data integration and the search and display capabilities that make GXD really shine. As of November 2024, for classical types of expression data, there are over 2 million annotated expression results for more than 16,000 genes with ~480,000 accompanying images.
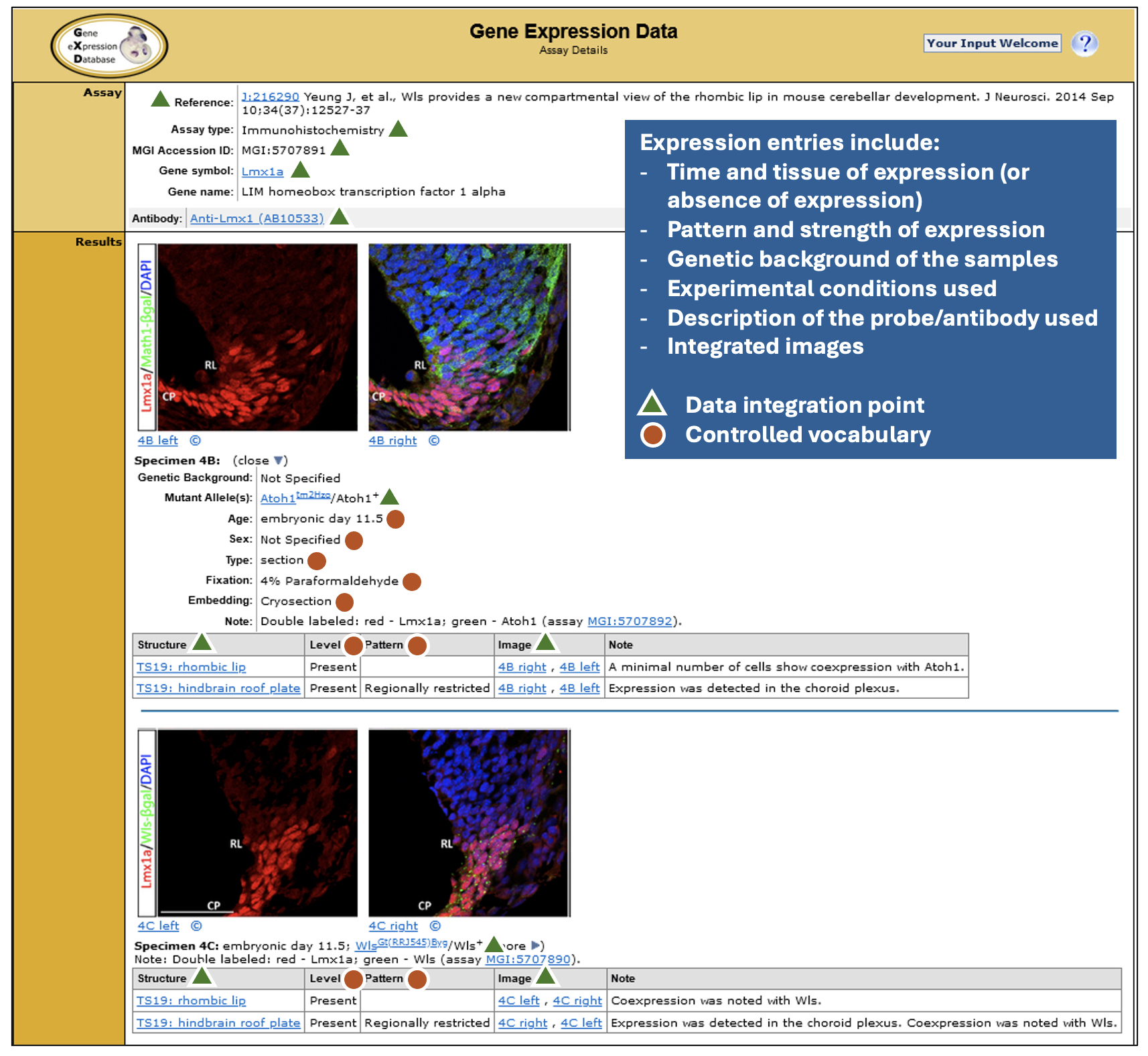
Index of publicly available RNA-seq and microarray experiments. More recently, we created an index of RNA-seq and microarray expression experiments deposited in GEO (Gene Expression Omnibus) and ArrayExpress. Finding experiments of interest in these repositories can be difficult because the entry metadata consists of free text provided by the submitters. To address this issue, we identify studies of endogenous gene expression in wild-type and mutant mice and annotate the experiment and sample metadata using the detailed controlled vocabularies and ontologies used elsewhere in GXD and MGI. Our current index of 8,000 mouse experiments includes standardized annotations for the anatomical structure, developmental stage, mutated gene, strain and sex of the samples, as well as the study type and key parameters of each experiment. Searches using this indexed metadata can be combined with free text searching of experiment titles and descriptions to allow you to find experiments of interest more effectively and reliably.
RNA-Seq expression data. GXD-relevant bulk RNA-seq data are imported from the European Bioinformatics Institute’s (EBI) Expression Atlas. The Expression Atlas generates uniformly processed TPM-level data sets for a select set of high-quality bulk RNA-seq experiments. As part of its incorporation into GXD, these data are further processed and annotated, resulting in seamless integration with GXD’s in situ and blot data.
How can researchers access data in GXD?
Search Forms. The GXD Home Page (http://www.informatics.jax.org/expression.shtml) provides the best entry point to all the features and resources provided by GXD. In addition to the Expression Literature and RNA-Seq and Microarray Experiment searches described above, of particular note are:
• Expression Data and Image Search – powered by GXD’s detailed annotations and data integration, this form provides the most fields, enabling basic or complex searches tailored to specific use cases.
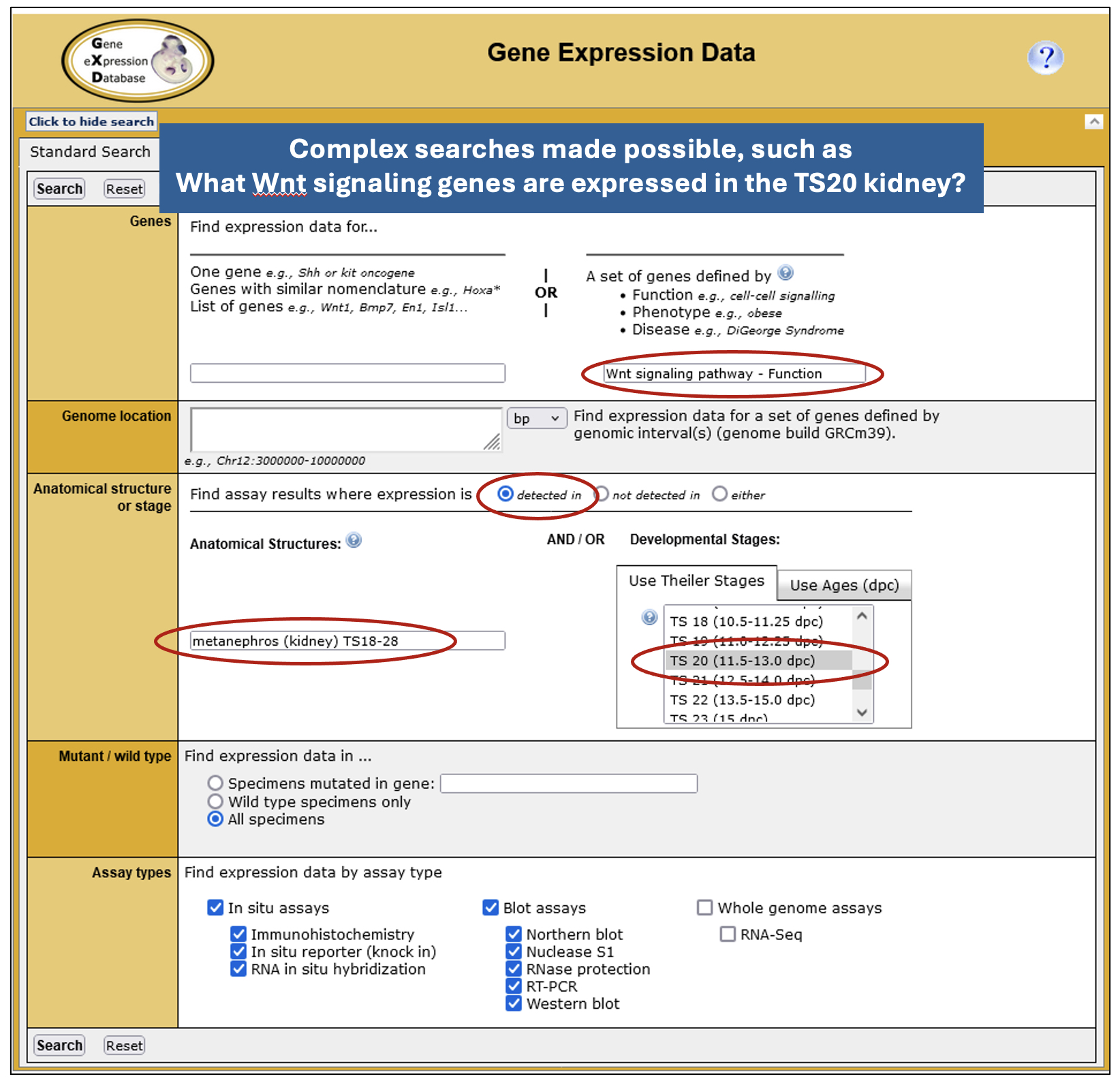
• Expression Profile Search – this allows you to search for genes by their expression profile. You can specify up to 10 anatomical structures and whether expression is present or absent in these structures. While currently limited to classical expression data, this search will soon be expanded to allow searching of RNA-seq data and the specification of developmental (Theiler) stages.
• Search Using Gene List – this permits you to retrieve GXD’s expression data for lists of genes and is, for example, useful for the further analysis of gene sets identified via high-throughput transcriptomic studies.
• Developmental Anatomy Browser – allows you to navigate through the extensive mouse developmental anatomy ontology used and maintained by GXD and provides links to the expression and phenotype data associated with those structures.
• MouseMine – provides programmatic access to GXD data.
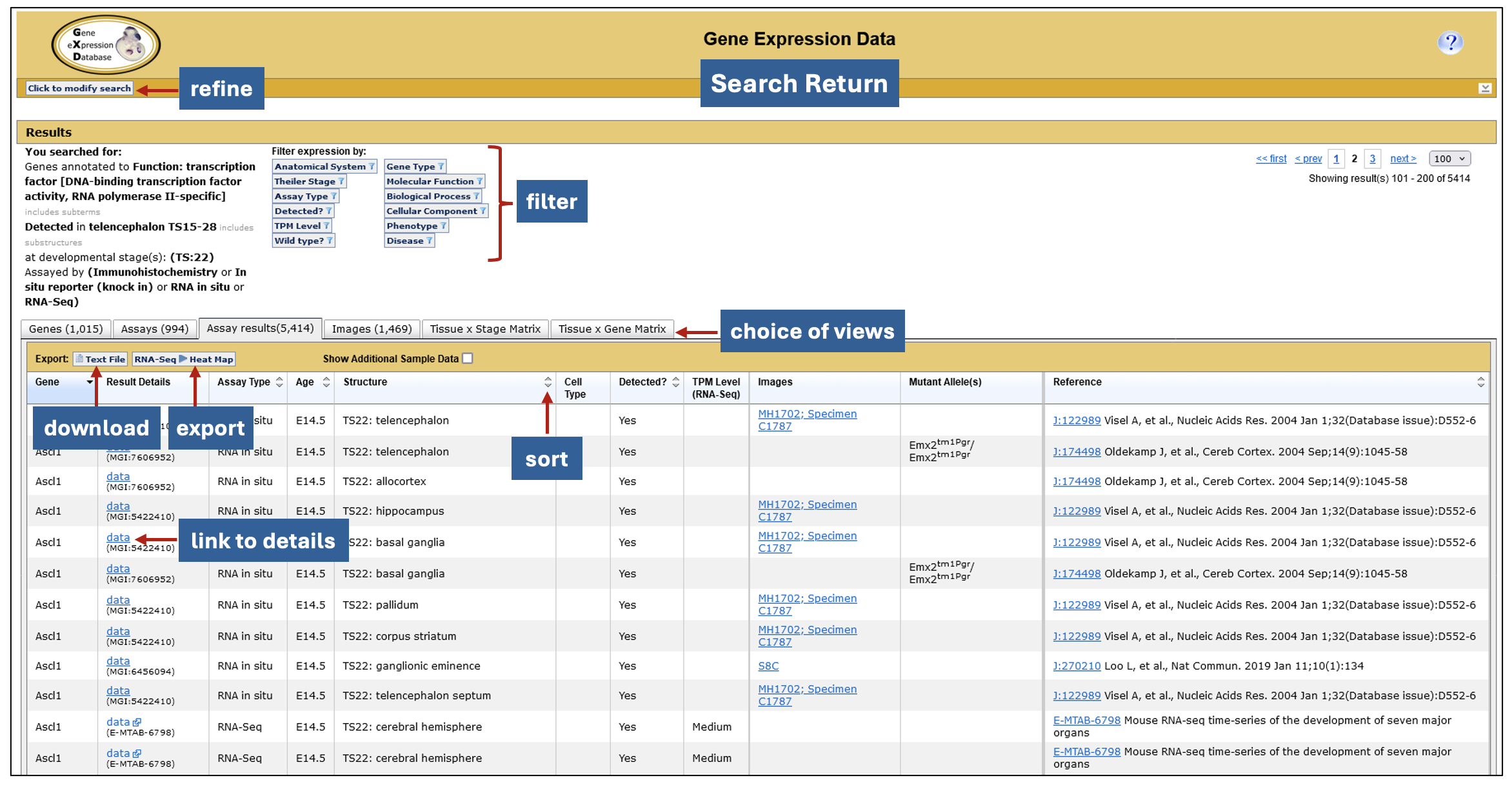
Interactive search returns. GXD expression data searches generate a six-tabbed data summary, allowing for different views of the search results. Filters that use the genetic, functional, phenotypic and disease-related information in MGI, as well as attributes of the returned expression data, allow you to tailor the return further. The data can be exported to other applications for further analysis including, in the case of RNA-seq data, Morpheus, a heat map visualization and analysis tool developed at the Broad Institute. Morpheus offers a myriad of utilities for further display and analysis, including sorting, filtering, hierarchical clustering, nearest neighbor analysis, and visual enrichment.
Any hidden gems that researchers might be less aware of?
Our gems are “hidden in plain sight” – the search forms described above. Most users of GXD/MGI use the Quick Search found in the upper right-hand corner of the pages. This provides a quick entry point into the data, but our search tools give you the capability to formulate queries using a wide variety of parameters derived from the integration of data in GXD and MGI. This allows you to successfully execute precision searches that can only be dreamt of at other resources.
Who are the people behind GXD?
GXD is located at The Jackson Laboratory in Bar Harbor, Maine. GXD scientific curators all hold Ph.Ds and have expertise in molecular and developmental biology. MGI has an integrated software team, supported by both the GXD and MGD grants, and integrated administrative and user support personnel. The following list represents our current staff. Many others have contributed to the success of GXD since 1993.
• Principal Investigator – Martin Ringwald
• Co-Principal Investigators (and co-leaders of the MGI Software Team) – Joel Richardson and Richard Baldarelli
• Scientific Curators – Jacqueline (Jackie) Finger, Terry Hayamizu, Ingeborg McCright, Constance (Connie) Smith, and Jingxia (Jing) Xu
• MGI Software Team – Jeffrey Campbell, Lori Corbani, and Pete Frost
• User Support – David Shaw
• Administrative Assistant – Janice Ormsby
Where does GXD’s funding come from?
GXD received startup funding from the W.M Keck Foundation. Since 1996 GXD has been supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD).
How can the community contribute to GXD?
We accept direct submissions. Review our Guidelines for submitting expression data and then contact us at gen@jax.org.
How can the community help GXD?
Cite GXD in your publications. This allows us to demonstrate our utility which helps us secure our funding.
What are the current and future directions of GXD?
We will be expanding our offerings to users who are interested in cell types. We have already begun to annotate expression data and samples from high-throughput expression experiments using terms from the Cell Ontology (CL). In the near future, users will be able to browse, search, and filter expression results and RNA-seq and microarray index entries by cell type.
We are also analyzing how we can represent and integrate results from newer types of expression analysis into GXD, namely single-cell RNA-Seq and spatial genomics data.
Following the plans outlined in our 1994 Science paper, we worked with the Edinburgh Mouse Atlas project (EMAP) to enable the spatial representation of expression data. Unfortunately, the EMAP project ended several years ago. However, similar projects and collaborations should hold big promise for dealing with spatial genomics data.
Where can we find GXD?
We are available at www.informatics.jax.org/expression.shtml.
References
Ringwald M, Baldock R, Bard J, Kaufman M, Eppig JT, Richardson JE, Nadeau JH, Davidson D. 1994. A Database for Mouse Development. Science 265:2033-2034. PubMed
Ringwald M, Mangan ME, Eppig JT, Kadin JA, Richardson JE, and the Gene Expression Database Group. 1999. GXD: A Gene Expression Database for the laboratory mouse. Nucleic Acids Res. 27:106-12. PubMed
Smith CM, Finger JH, Hayamizu TF, McCright JJ, Eppig JT, Kadin JA, Richardson JE, and Ringwald M. 2007. The mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res. 35: D618-D623. PubMed
Finger JH, Smith CM, Hayamizu TF, McCright IJ, Xu J, Eppig JT, Kadin JA, Richardson JE, Ringwald M. 2015. The mouse gene expression database: New features and how to use them effectively. Genesis. doi: 10.1002/dvg.22864. PubMed
Hayamizu TF, Baldock RA, Ringwald M. 2015. Mouse anatomy ontologies: enhancements and tools for exploring and integrating biomedical data. Mamm Genome. 2015 Oct;26(9-10):422-30. PubMed
Finger JH, Smith CM, Hayamizu TF, McCright IJ, Xu J, Law M, Shaw DR, Baldarelli RM, Beal JS, Blodgett O, Campbell JW, Corbani LE, Lewis JR, Forthofer KL, Frost PJ, Giannatto SC, Hutchins LN, Miers DB, Motenko H, Stone KR, Eppig JT, Kadin JA, Richardson JE, Ringwald M. 2017. The mouse Gene Expression Database (GXD): 2017 update. Nucleic Acids Res. 2017 Jan. 4;45 (D1): D730-D736. PubMed
Smith CM, Hayamizu TF, Finger JH, Bello SM, McCright IJ, Xu J, Baldarelli RM, Beal JS, Campbell JW, Corbani LE, Frost PJ, Lewis, JR, Giannatto SC, Miers DB, Shaw DR, Kadin JA, Richardson JE, Smith CL, Ringwald M. 2019. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019 Jan. 8;47 (D1): D774–D779. PubMed
Smith CM, Kadin JA, Baldarelli RM, Beal JS, Blodgett O, Giannatto SC, Richardson JE, Ringwald M 2020. GXD’s RNA-Seq and Microarray Experiment Search: using curated metadata to reliably find mouse expression studies of interest. Database 2020 Mar. 4 PubMed
Baldarelli RM, Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Shaw DR, Beal JS, Blodgett O, Campbell J, Corbani LE, Frost PJ, Giannatto SC, Miers DB, Kadin JA, Richardson JE, Ringwald M 2021. The mouse Gene Expression Database (GXD): 2021 update. Nucleic Acids Res. 2021 Jan 8;49(D1):D924-D931. PubMed
Ringwald M, Richardson JE, Baldarelli RM, Blake JA, Kadin JA, Smith CL, Bult CJ. 2021. Mouse Genome Informatics (MGI): latest news from MGD and GXD. Mamm Genome. Oct 26, 2021 PubMed


 (1 votes)
(1 votes)