Featured Resource: Xenbase
Posted by the Node, on 7 December 2023
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. Our ‘Featured resource’ series aims to shine a light on these unsung heroes of the science world. In this post, the team behind Xenbase introduces the key features on the database and suggests how the community can contribute to the mission of Xenbase.
When was the Xenbase established and what are its aims?
Xenbase was the brainchild of Peter Vize. Originally conceived of as an online catalog of gene expression images in the late 1990’s, the golden age of gene expression screens, where essentially every lab was producing 100s of images of gene expression throughout embryonic development, yet those pictures were sitting on a lab computer not being shared or annotated in any meaningful way. Peter saw that a resource to share this information would have a huge impact to cut down on wasted time, effort and resources (i.e. save funding dollars).
The first iteration of Xenbase was launched in 2000, and by 2002, Xenopus tropicalis was earmarked for whole genome sequencing by the Joint Genome Institute (JGI), so the vision for Xenbase quickly morphed into a bigger project: integrated genomics and gene expression on a fully searchable database.
The initial challenge faced when building Xenbase was to combine the research from two Xenopus species used in complementary but (almost) non-overlapping fields of cell biology and embryology. Xenopus laevis had a long history as a lab frog, with decades of literature covering organogenesis, cell fate maps and cell biology, gene function and gene expression, but as a polyploid of hybrid origin, the genome hadn’t been sequenced (and was a long way off). In comparison, the diploid Xenopus tropicalis genome was being sequenced and this smaller frog was being adopted for disease modeling, and had lots of EST data, but there wasn’t a lot of other biological data for ‘trops’. It was clear both Xenopus species were in need of database support, and the Xenbase founders met that challenge head on, in large part by learning from the already established MODs like MGI (mouse) and Zfin (zebrafish). Xenbase was the first MOD to support two species and essentially three genomes (i.e., X. tropicalis and the 2 subgenomes of X. laevis).
The overarching aim of Xenbase is to simultaneously support labs using Xenopus as a research model, share the genomes and bioinformatic information about genes/proteins, codify the results of the research via deep and expert curation and thus support basic and applied science to accelerate discovery. Having all the data about Xenopus in one place has huge advantages. Within a few years enough people found that ‘what works for X. laevis, works for X. tropicalis’, e.g., gene expression in X. laevis and X. tropicalis are nearly always the same (or very similar), so the same reagents (such as MOs, gRNAs, and antibodies) can be used in both species.
Who are the people behind the resource/ who runs the resource?
Xenbase has two teams and two performance sites. The curation team is based at the Division of Developmental Biology, at Cincinnati Children’s in Ohio, USA, led by Prof. Aaron Zorn. We currently have four curators and bioinformatician/genome analyst in the Cincinnati-based curation team: VG Ponferrada, Malcolm Fisher, Andrew Bell, Christina James-Zorn, and Ngoc Ly. We also have a student assistant, Nguyen Thuy Vy Ngo, who helps triage the new literature. The development team, headed by Prof. Peter Vize, is based in the Department of Biological Sciences at the University of Calgary, in Alberta, Canada. The Calgary development team is led by Kamran Karimi, with support of the database architect Troy Pells, and the Bioinformatian/genome specialist, Vaneet Lotay, who are further supported by software developers, Joe Wang and Stan Chu. We also share code and development with our sister-website, Echinobase, so the Calgary based team is ably supported by Brad Arshinoff and Sergei Agalakov.
What tools/resources are available for researchers?
Xenbase is a gene-centric database, with a single ‘umbrella’ gene page showing the X. tropicalis gene and the two X. laevis paralogs, typical gene expression images at different embryonic stages (when available) and other data such as reagents, orthologs and OMIM/DO associated diseases (and many more links to associated data). Each gene page then has a series of tabs, like a folder, starting with the Expression (all images in database with expression), Phenotypes (all experiments that either manipulate or assay the gene(s)) and Literature (all literature that cite the gene(s)). The next set of tabs cover GO terms, Nucleotides, Proteins and Interactants, which collate annotations and/or accession about the genes/protein products with data pulled in from, and linked to, numerous trusted databases (NCBI, ENSEMBL, UniProt, InterPro/TrEMBL, GO and IntAct).
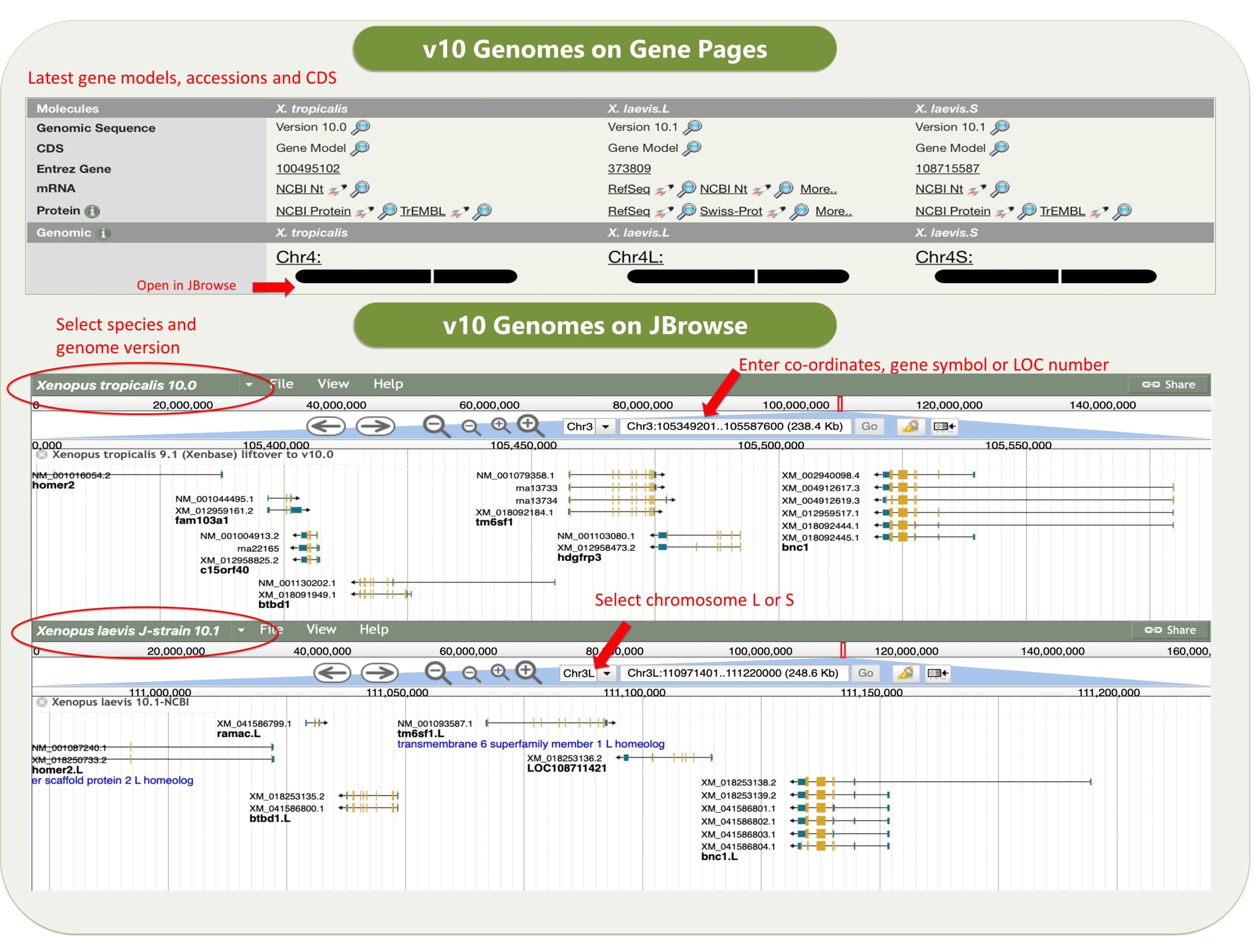
Xenbase has various tools that are widely used. The genome viewer JBrowse has the latest v10 Xenopus genomes, and a huge variety of other useful tracks such as CRISPR-Scan predicted guide RNAs to help design your mutant lines, a morpholino track, and an enormous number of RNA-seq and ChIP-seq tracks, tracks for histone marks and transcriptions factor binding sites, and many more.
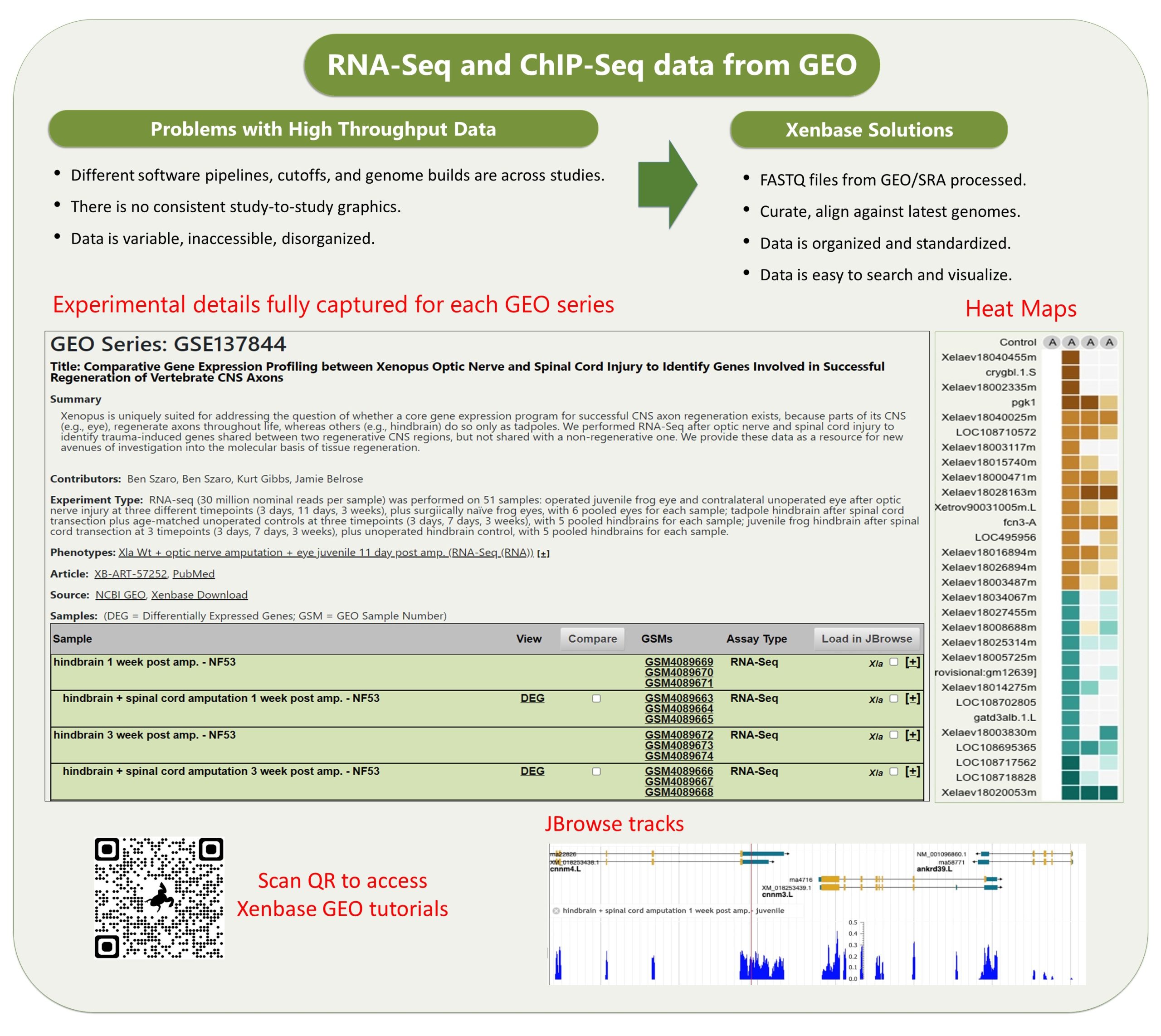
The other data module which is really cool is our GEO data ( accessed via Expression Menu/GEO data @ Xenbase). We took the publicly submitted high throughput sequence data from the NCBI’s GEO database, manually curated the supporting metadata and then we processed it through a pipeline that ‘harmonizes’ the different studies (see Fortriede et al 2020 for all the details). Even though the data are from different studies, researchers can view all of this data in a standardized format, aligned to the latest v10 genomes and via heat maps that visualize DEGs (differentially expressed genes). In addition, we have 1000’s of ‘computational’ gene expression phenotype statements from these experiments, where we generate statements in the readable format, e.g., “manipulation X increases/decreases the expression of gene Y in tissue Z at NF stage #”, all linked to genes, literature and the original GEO data. These ‘gene expression phenotypes’ are most easily returned via our Phenotypes search (e.g., search for six1 . We hope this mass of curated RANSeq and ChIPSeq data will help inform GRNs, and let researchers see all the results from other peoples experiments that pertain to their gene of interest.
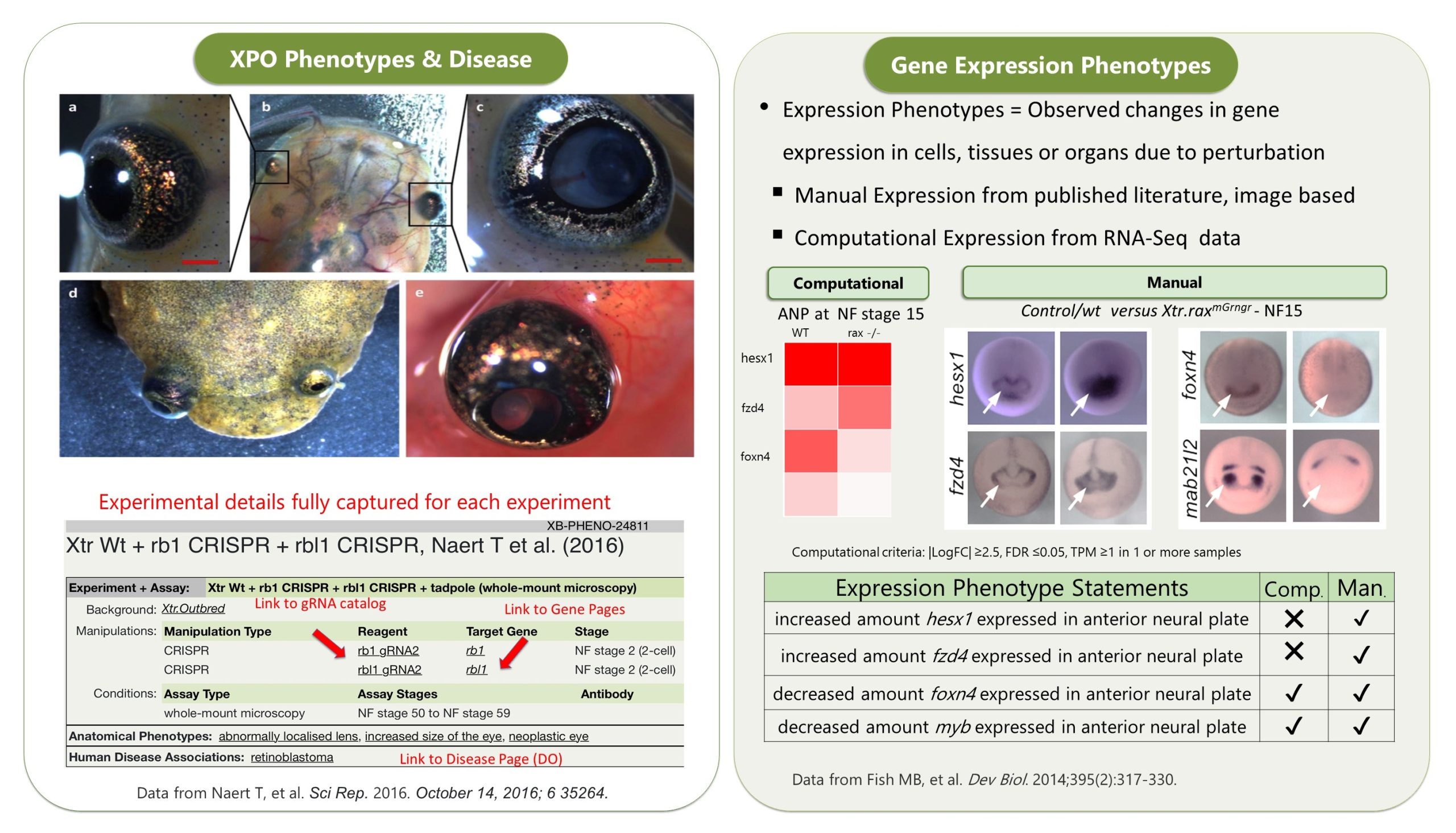
Any hidden gems, features that are new, or that researchers might be less aware of?
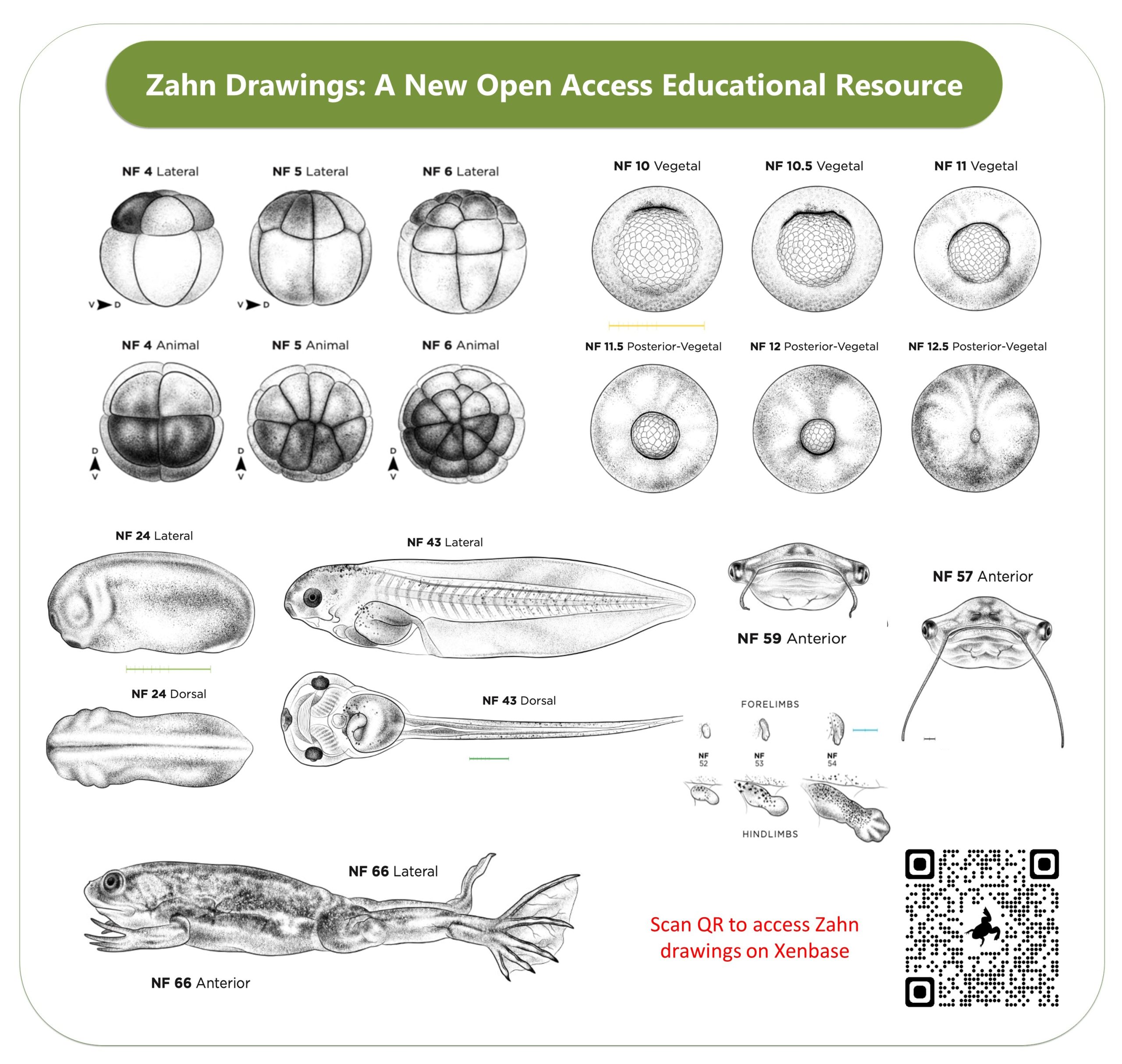
Two new features we are super proud of are the open access drawings of Xenopus embryonic stages (the Normal table) by Natalya Zahn and the accompanying Landmarks Table. The Zahn drawings, which are in the same style as the classic 1950’s Nieuwkoop and Faber drawings, are open access and cover more views- especially anterior and ventral views. Also, the accompanying Staging Landmarks Table that we built to help researchers in the wet lab stage embryos also includes internal developmental milestones and gene markers. We hope both of these resources become indispensable for university courses, in wet labs and embryology courses.
I think the Xenopus Community pages, which include personal profiles and lab pages, are both great ways to promote one’s research and attract students and collaborators. I recommend PIs appoint a trainee in the lab to make sure their Xenbase profiles & Lab page is up-to-date, and that all members of the Lab have a profile to record their research interests and a list of their publications. The Xenbase Jobs Board is also available to post any open positions, from graduate students to postdocs and group leaders and department chairs.
We are constantly working to keep the information on Xenbase up to date and synchronized with the other major databases and repositories, and we recently joined the Alliance of Genome Resources. By collating data from diverse model organisms (worm, yeast, fly, mice, frogs, rats and fish) the Alliance aims to improve the understanding of the genetic and genomic basis of human biology, health and disease. Frogs have played, and continue to play, an important part of that discovery process.
How can the community contribute?
The single most effective way for researchers to contribute to the mission of Xenbase, is to choose open access (OA) options to publish their research, whenever possible. When data is locked up behind a paywall, we literally can’t see it, so we can’t curate it. If we don’t curate the data, the results become essentially invisible over time, and its immediate and long term impact is greatly reduced. Accessible data becomes curated data, which is discoverable and will be cited more too. Uncurated data is easily overlooked.
Another way people can contribute is to send us their images of novel gene expression, especially images of stages beyond what is included in their papers. Instead of being saved in an inaccessible folder on a lab computer, send them to Xenbase and share them with the world!. All community submitted images are fully attributed to the people and that lab that makes them!
We also run a help desk: xenbase@cchmc.org. Contact us anytime with community submissions, questions, feedback, concerns, troubleshooting help and your ideas to make Xenbase an even better, more useful resource.
Where does funding come from?
Xenbase is currently supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). Previous financial support also came from the National Science Foundation (NSF), the Wellcome Trust (UK), the BBSRC (UK), the Alberta Network for Proteomics Innovation (ANPI).
If money was no object, what would you like to add to Xenbase?
We’re looking into some new great tools including updating to JBrowse2 (to allow viewing multiple genomes at once), and new data graphics to show/assess synteny across Xenopus genomes, and supporting single-cell data. We’d also like to develop more educational resources to support students and teaching labs, including a high tech histology or a 3D atlas of Xenopus from embryos to adults, more anatomy atlas modules, perhaps including virtual dissection of adult Xenopus frogs. We would like to fund more illustrations of Xenopus development, to really fill in the Normal Table we published in Zahn et al 2022 in Development! We would also love to get movies (which are more and more common in publications) embedded on the articles pages. All of these ideas have been floated, so we’ll see what we can get done.
References
Fortriede JD, Pells TJ, Chu S, Chaturvedi P, Wang D, Fisher ME, James-Zorn C, Wang Y, Nenni MJ, Burns KA, Lotay VS, Ponferrada VG, Karimi K, Zorn AM, Vize PD, Xenbase: deep integration of GEO & SRA RNA-seq and ChIP-seq data in a model organism database, Nucleic Acids Res., Volume 48, Issue D1, Pages D776-D782, doi:10.1093/nar/gkz933
Zahn N, James-Zorn C, Ponferrada VG, Adams DS, Grzymkowski J, Buchholz DR, Nascone-Yoder NM, Horb M, Moody SA, Vize PD, Zorn AM, Normal Table of Xenopus development: a new graphical resource, Development, 2022 Jul 15;149(14):dev200356, doi:10.1242/dev.200356
Fisher M, James-Zorn C, Ponferrada V, Bell AJ, Sundararaj N, Segerdell E, Chaturvedi P, Bayyari N, Chu S, Pells T, Lotay V, Agalakov S, Wang DZ, Arshinoff BI, Foley S, Karimi K, Vize PD, Zorn AM. Xenbase: key features and resources of the Xenopus model organism knowledgebase. Genetics. 2023 May 4;224(1):iyad018. doi: 10.1093/genetics/iyad018. PMID: 36755307; PMCID: PMC10158840.


 (1 votes)
(1 votes)