Geometrical models of tissue as tools for uncovering rules underlying tissue organization
Posted by Hernán Andrés Morales-Navarrete, on 27 May 2016
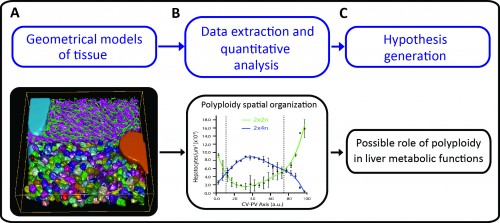
A major challenge in cell and developmental biology is to understand the mechanisms whereby cells interact with each other to form the variety of complex tissue forms present in organisms. This requires visualizing and analysing different cellular processes across multiple scale levels -from the subcellular to the tissue, i.e. generating cell and tissue models at different levels of complexity. The different cells as well as their essential sub-cellular components can be observed by fluorescence microscopy and reconstructed by means of image processing techniques. Then, three-dimensional (3D) digital representations of tissue can be generated using the structural information extracted from the microscopy images. A set of 3D digital representations of tissue at multiple scale levels constitutes a geometrical model of the tissue. Such a geometrical model provides the means of extracting detailed morphological and structural information of the tissue as well as its spatial variability. Based on such information, new hypotheses about tissue organization can be generated (Figure 1).
In our recent study published in eLife, we developed a software platform for the reconstruction and quantitative analysis of tissue architecture in 3D (Morales-Navarrete et al., 2015). We combined newly developed and established image analysis algorithms in a pipeline that allows for a flexible workflow to reconstruct and analyse different structures forming the tissue with high accuracy. It is implemented as part of the stand-alone freely available software MotionTracking (http://motiontracking.mpi-cbg.de; the name is historical as it was first developed for the tracking of endosomes in cultured cells (Rink et al., 2005)) and addresses several unmet computational needs, such as 1) high accuracy of 3D geometric reconstruction, 2) image processing of large volumes of tissue, 3) fast image analysis amenable to middle-throughput, 4) it can be run on a regular PC, 5) it is adaptable to various tissues and imaging conditions, 6) it provides a flexible tool for tissue reconstruction without need of further programming or scripting and 7) it allows quantitative morphometric and spatial analysis at cellular and tissue level.
As proof of principle, the pipeline was applied to the analysis of liver tissue of adult mice. The resulting geometrical model provided a complete description of different morphological parameters across multiple scales from tissue to subcellular level (Figure 1A and http://dx.doi.org/10.7554/eLife.11214.003, http://dx.doi.org/10.7554/eLife.11214.012, http://dx.doi.org/10.7554/eLife.11214.022) and uncovered a significant amount of new biological information. For example, we could map the distribution of different populations of hepatocytes with different number of nuclei and DNA content within the mouse liver lobule. It has been reported that hepatocytes are remarkably heterogeneous in terms of ploidy but whether they are randomly distributed or follow a specific spatial organization remains controversial (Gentric and Desdouets, 2014). Our quantitative and spatial analysis of the geometrical model revealed an unexpected spatial distribution of hepatocytes with distinct DNA content. Whereas hepatocytes with low ploidy were enriched in the peri-central and peri-portal regions, high ploidy hepatocytes were preferentially located in the middle region (Figure 1B and http://dx.doi.org/10.7554/eLife.11214.027). This is the first time that polyploidy is found to be zonated and follow a specific pattern.
Such an unexpected zonation suggests the existence of mechanisms underlying the functional specialization of hepatocytes during tissue homeostasis. One of the most remarkable features of liver tissue is the zonation of metabolic functions (hepatocytes located in the vicinity of the portal vein have different metabolic activities than hepatocytes located near the central vein). Our findings therefore suggest a possible role of polyploidy in liver metabolic functions (Figure 1C). Moreover, low ploidy hepatocytes (enriched in the peri-central and peri-portal regions) may correspond to the cells recently implicated in hepatocyte renewal (Wang et al., 2015, Font-Burgada et al., 2015). Altogether, these results show how tissue geometrical models can be used to unravel principles of tissue architecture but also generate hypotheses about tissue structure and function.
We expect our pipeline to be very useful to the biology community since it can be used for several applications from basic (understanding of how cells form tissues) to clinical research (diagnosis of diseases by 3D digital histology).
Software resources:
All resources for our platform, including user manuals, examples and the latest version of the software, can be found on http://motiontracking.mpi-cbg.de/get/.
Main Reference:
MORALES-NAVARRETE, H., SEGOVIA-MIRANDA, F., KLUKOWSKI, P., MEYER, K., NONAKA, H., MARSICO, G., CHERNYKH, M., KALAIDZIDIS, A., ZERIAL, M. & KALAIDZIDIS, Y. 2015. A versatile pipeline for the multi-scale digital reconstruction and quantitative analysis of 3D tissue architecture. Elife, 4.
Other References:
FONT-BURGADA, J., SHALAPOUR, S., RAMASWAMY, S., HSUEH, B., ROSSELL, D., UMEMURA, A., TANIGUCHI, K., NAKAGAWA, H., VALASEK, M. A., YE, L., KOPP, J. L., SANDER, M., CARTER, H., DEISSEROTH, K., VERMA, I. M. & KARIN, M. 2015. Hybrid Periportal Hepatocytes Regenerate the Injured Liver without Giving Rise to Cancer. Cell, 162, 766-79.
GENTRIC, G. & DESDOUETS, C. 2014. Polyploidization in liver tissue. Am J Pathol, 184, 322-31.
RINK, J., GHIGO, E., KALAIDZIDIS, Y. & ZERIAL, M. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell, 122, 735-49.
WANG, B., ZHAO, L., FISH, M., LOGAN, C. Y. & NUSSE, R. 2015. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature, 524, 180-5.


 (3 votes)
(3 votes)