Grooming a new generation of technology savvy scientists at NMMP2022 Summer School in Transcriptomics
Posted by Valeria Ghezzi, on 18 July 2022
Lenka Belicova, Valeria Ghezzi.
Where Science meets Technology – NMMP2022 Summer School in Transcriptomics

In Sweden, where the sun doesn’t go down in the summer, the science stays awake!
“57 students, 20 universities and research institutes, 24 nationalities and 1 common interest: learning more about omics approaches.”: tweeted Professor Silvia Remeseiro, which together with Professor Claudio Cantù and Professor Andreas Moor co-organized the first ever NMMP2022 Summer School in Transcriptomics during Development and Cancer in an amazing location of Sandvik Gård on June 27th-30th, 2022.
One of the intriguing features of the Summer School was a relatively low ratio between students and teachers. Ten top-notch scientists coming from all over Europe and beyond helped students dive into a broad range of fascinating topics from newest technologies in single cell transcriptomics to 3D organization of chromatin.
Science unlimited
The four-day program of the Summer School was packed with exciting lectures by keynote and invited speakers, insightful “meet the speaker” sessions providing a glimpse on the life of scientists, and opportunities to discuss students’ projects and questions that are driving them. The Summer School succeeded in bringing together scientists at different career stages from variety of fields inspiring one another.

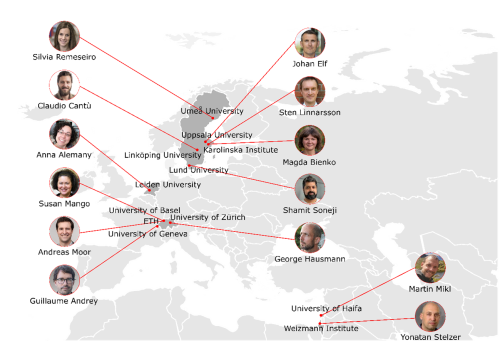
Students had the chance to listen to amazing lectures given by keynote speakers Prof. Sten Linnarsson, Prof. Johan Elf and Prof. Susan Mango, as well as the ones given by the invited speakers: Dr. Yonatan Stelzer, Prof. Anna Alemany, Dr. Shamit Soneji, Prof. Martin Mikl, Prof. Guillaume Andrey, Prof. Magda Bienko and Dr. George Hausmann.
Read more about about the speakers and organisers in the “Meet the speakers” paragraph later on this page!
Blurring the lines between dry and wet
Many biological questions require computational approaches to be explored in depth. Experimentalists have to find a common language with bioinformaticians and computational scientists and vice versa, which is not always an easy task. The organizers know it well and designed the program with the hope to generate ample opportunities for “wet” and “dry” scientists to exchange ideas and discuss how to build a bridge between these two domains. Indeed, according to the poll (Fig. 3), the participants had diverse experiences from “dry” and “wet” lab, and almost a fifth was open to transition.
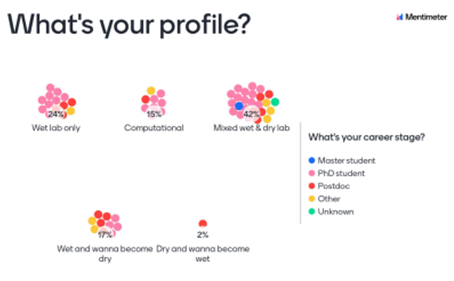
One of the ways to find a common language is to try to explain our projects to researchers outside of our immediate niche. The students tried this during a new poster session format: the “Non-poster” poster session. For two and a half hours, the students divided into small groups presented their posters and provided feedback to each other. The fun part: the projects were as different as it gets. The students left the poster session energized, inspired and willing to try new approaches to their problems. The only complaint they had: “We were enjoying it too much! Two hours and a half were not enough!”. We are sure organizers will take a note for the next edition of the Summer School.

Picture taken by Professor Claudio Cantù.
A dive into data
The science at the Summer School was at the top of innovation. For example, students got to hear about a new, fresh from the press, scRNA-seq technology VASAsequencing by Prof. Anna Alemany. Many students presented their projects on developing the technology of the future we are looking forward to putting in practice one day.
Perhaps one of the most immersive experiences of the future of transcriptomics data analysis was provided by Dr. Shamit Soneji and his team that developed CellexalVR software. Using virtual reality, students were able to literally dive into single-cell data and visualize the data with a whole new perspective!

Picture taken by Dr. Pierfrancesco Pagella.
Technology has its place in biological research
One could argue that recent fancy technological developments push us paradoxically further away from answering biological questions. Recently, this topic ignited a vivid debate on the Node platform and Twitter. The Summer School was a perfect place to continue discussing this topic as it united biologists experimentalists on one hand and technology fans on the other. Prof. Magda Bienko, avid fan of technological advances and pioneer in understanding the 3D organization of chromatin, added an interesting angle to the debate. We should give enough time for the technology to be developed fully, so all its caveats are addressed before we use it to answer a biological question. If we do it too early, we risk overinterpretation of the results, confusion and mistrust in technology. Technology has a great promise to help us understand the world around us, but we should not rush the process and trust that it will deliver once optimized properly.
Consequently, we witnessed a couple of “conversions” of pure experimentalists willing to give a new technology a try: “Before the Summer School, I saw technology as something that brings me away from the lab work, losing contact with the magic of seeing biology unfolding in front of your eyes. But I realized that technology is just a wonderful mean to get a deeper insight into biological mechanisms. Now I can’t wait to go back and put my hands on one of these amazing tools I learned about here!” stated one of the early-career-stage students.
Such realizations don’t come as a surprise given the keynote and invited speakers are leaders in the leveraging technological advances to gain insights into biology and elicited great enthusiasm in all of the
students present at the Summer School with their talks.
Meet the speakers
- Prof. Sten Linnarsson’s work is shaping the transcriptomics field and transforming our view of the multitude of cells type that emerge during animal development. With his group, he crafted new ways to identify secret messages hidden within transcriptomics datasets.
- The work of Prof. Johan Elf is bringing the gene regulation field into a new dimension, as it is producing quantitative physical models and biological observations that, when merged, enable to determinate the real behavior of molecules.
- The contributions of Prof. Susan Mango and her group are incredible: from identifying master regulators that shape cell fate, to the compelling and almost counteracting notion that variable environmental conditions play a non-negligible role on developmental trajectories. She is transforming C. elegans into a star of developmental biology.
- Dr. Yonatan Stelzer’s group is implementing and developing cutting-edge genome- and epigenomeediting tools together with sophisticated epigenetic and gene expression reporters on embryonic stem cells and developing mice.
- Prof. Anna Alemany‘s training and scientific production position her work among the emerging stars in the field of single-cell transcription, and how this can be used to understand cell-fate commitment. She contributed to developing a new scRNA-seq technology – VASAseq – that gives high throughput
full gene body read-out with single cell resolution. - Dr. Shamit Soneji’s group is developing analytical pipelines in a new dimension: they recently developed Virtual Reality tools to navigate and analyse most sophisticated datasets. Among these tools, CellexalVR is a platform for visualisation and analysis of single-cell gene expression data.
- Prof. Martin Mikl works on generating new ways to identify, in a high-throughput and standardized manner, the rules that govern mRNA function and localization.
- Prof. Guillaume Andrey is an emerging leader in the field of the 3D genome, he focuses on the ideological and conceptual innovation of how the genome could be regulated in 3D in addition to the DNA sequence.
- Prof. Magda Bienko developed GPseq: a new creative technique that allows mapping the radial organization of the human genome, revealing new patterns of genomic and epigenomic features, gene expression, and activity compartmentalization.
- Dr. George Hausmann has an enviable scientific experience and writing dexterity: the entire Department of Molecular Life Science in Zurich competes with his time for help or consultation on manuscript design, writing and perfectionism.
Organizers:
- Prof. Claudio Cantù’s Lab, at Linköping University, is focused on discovering the mechanisms of genome regulation that drive specialization during embryonic development using sophisticated tools, from mouse genetics to high-throughput state-of-the-art biochemical approaches. The group is focused on the so-called ‘Wnt signalling pathway’, a molecular cascade important for virtually all aspects of development, and whose deregulation causes human malformations and several forms of aggressive cancers.
- Prof. Silvia Remeseiro works as a Wallenberg Molecular Medicine Fellow in Umeå University. Her group is mainly focused on how the reprogramming of regulatory regions and topological changes in 3D chromatin organization determine gene dysregulation in glioblastoma, and how this subsequently contributes to malignancy, heterogeneity and invasiveness.
- Prof. Andreas Moor works at the Department of Biosystems Science and Engineer, at ETH Zurich, focusing on exploring the way in which single cells collaborate within tissues to achieve their common functions. His group makes use of quantitative approaches to study cellular and subcellular heterogeneity while preserving information about the spatial tissue context.
Future is in good hands
The Summer School was a dream come true for the organizers Claudio Cantù, Silvia Remeseiro and Andreas Moor, finally taking place after the delay caused by the pandemic. Their hope was to provide similar experiences, that shaped their scientific thinking when they were trainees, to a next generation.
Probably, even their wild imagination was exceeded by the success of this edition of the Summer School: 56 students from all over Europe and one, Vasikar Murugapoopathy, from McGill University in Canada, thanks to a travel grant offered by Antibodies online GmbH.

High quality of student talks, their engagement and drive to discuss science till late hours impressed the tutors.
When Prof. Susan Mango was asked to describe the Summer School in one word, she answered: “My word would have been STUDENTS. They were great – very interactive, smart questions. A really good lot”.
Prof. Anna Alemany later revealed: “It was really motivating, from the speaker point of view, to discuss the different aspects of science during lunch, coffee breaks, and walks around the lake. The motivation was contagious, I am sure this group of people will achieve anything they want!”.
It looks like the future of life science and molecular medicine is in good hands.
Picture taken by Prof. Claudio Cantù.
Shall we meet next year?
The Summer School was generously supported by a collaborative grant of The National molecular Medicine Fellows Program (NMMP) in Sweden awarded to the organizers Claudio Cantù and Silvia Remeseiro.
Both are part of NMMP network: 100 group leaders recruited to the Wallenberg Centers of Molecular Medicine, which is co-funded by SciLifeLab and Knut and Alice Wallenberg Foundation.
BioNordika and Antibodies online GmbH were the only two, carefully chosen additional sponsors.
Dr. Stefan Pellenz, accomplished scientist and currently product manager at Antibodies online GmbH,
shared his insights on how to profile the epigenome with CUT&RUN and CUT&Tag methods.
We hope the Summer School was not a one-time experience and organizers will be able to get support for the next edition.
With an amazing venue, new connections created, excitement shared about the future of science, participant enjoyed the Summer School so much that they unanimously voted for the next edition!
Want to be part of the future? Join the next edition on NMMP2022 Summer School in Transcriptomics!
Get in touch with us:
Wallenberg Centre For Molecular Medicine (Linköping)
Knut och Alice Wallenbergs Stiftelse
National Molecular Medicine Programme
HELP US SPREAD THE VOICE! #NMMPschool2022




 (2 votes)
(2 votes)
I am so glad to seeing this post. It is a wonderful description of the event by Lenka Belicova and Valeria Ghezzi.
I can add that, during this summer school, the PIs have learnt a lot from the incredible students who attended.
Thank you!
Claudio