Hox genes: the key to decipher limb position – the story behind the paper
Posted by Chloe Moreau, on 6 February 2019
In our recent paper published in Current Biology, we unravel the direct and early role for Hox genes in the regulation and natural variation of the forelimb position in birds. Here I will share with you the story behind this paper.
I have always been fascinated by the question of how, from a single cell, a multicellular organism with a complex, highly organized three-dimensional structure can arise. Therefore, when I started my PhD in Dr. J. Gros’ lab and got the opportunity to work on the question of limb position in chicken embryos, I was thrilled. How limbs reproducibly form along the vertebrate body is a fascinating, long standing question for developmental but also for evolutionary biologists. Indeed, this question has two main aspects:
- one purely developmental aspect – in trying to understand the cellular and molecular mechanisms establishing limb position during the embryo development
- one evolutionary aspect – in tackling the question of diversity in limb position amongst tetrapod species.
The context
In tetrapods, while limbs always position at the level of the cervico-thoracic (for the forelimb) or lumbo-sacral (for the hindlimb) vertebral transitions, the position of these vertebral frontiers – and hence the limb position – is highly variable between species. Avian species especially, display a wide variation in the position of their forelimb – from the sparrow that form forelimbs at the level of the 10th vertebra up to the swan in which forelimbs form at the level of the 25th vertebra – a difference of no less than 15 vertebrae!
Despite major advances in our understanding of limb patterning in three dimensions, how limbs reproducibly form along the anteroposterior axis and how variations in these positions arise remained largely unresolved. Hox genes were long suspected to regulate limb position (Tanaka, 2013; Tickle, 2015). This assumption first arose because of their well-demonstrated role in patterning the vertebrae and was further supported by cross-species comparative studies which showed that Hox gene expression domains correlate with limb position in different species (Burke et al., 1995). But the different supportive evidences were mostly correlative (Burke et al., 1995; Cohn et al., 1997; Minguillon et al., 2012; Nishimoto et al., 2014). In addition, whereas Hox gene mutant mice display vertebral identity transformations (Mallo et al., 2010), functional studies in support for a role of Hox genes in regulating limb position were lacking. Therefore, whether Hox genes would control limb initiation and position was clearly unresolved at the time we started this work.
Our Results
Where do the limbs come from? They originate from the Lateral Plate Mesoderm (LPM), a mesodermal tissue that flanks axial embryonic structures (i.e. notochord, neural tube and somites), and will emerge from this tissue at their stereotypical position, at three days of development (Hamburger and Hamilton stage 15). First, we wanted to determine when the forelimb position is primarily established. We took advantage of the possibility to do grafting experiments in chicken and quail embryos, combined with the use of transgenic quail lines generated in the lab, to establish that the forelimb position is determined very early, 24h before limb initiation (i.e. at stage 11, 2 days of development).
The finding that the forelimb position is already established by stage 11 led us to think that this positional information could be established earlier – during the process of gastrulation. Gastrulation is this key morphological process during which the three germ layers – ectoderm, mesoderm and endoderm – are formed. The LPM is generated during this process and we wondered whether it was also patterned into limb- and non-limb domains while being generated. Whereas LPM precursor cells in the epiblast had been identified through lineage-tracing experiments (Psychoyos and Stern, 1996), how the forelimb, interlimb and hindlimb cells are generated was not characterized. The process of gastrulation is really dynamic and spans for about 24h. Therefore, in order to precisely catch the dynamic behaviors of LPM precursor cells, we used yet again another advantage of the chicken embryo, which is the possibility to do live-imaging, especially in the early stages of development as the embryo is flat and can be cultured ex ovo. We therefore performed a dynamic lineage analysis of the LPM formation, and could nicely characterize that the forelimb, interlimb and hindlimb domains are sequentially generated during gastrulation.
At this point, we had described how the LPM is formed and patterned into limb and non-limb domains at the cellular level. The next step was now to investigate the molecular mechanism underlying this process.
As I mentioned earlier, our top candidates for the regulation of limb position were the Hox genes, well-known for their role in patterning vertebrae along the main body axis (Mallo et al., 2010). For our study, it is important to remember that these genes are arranged in four different clusters and display a specific chromosomal organization that reflects their sequence of activation (i.e. temporal collinearity) and their successive domains of expression along the antero-posterior axis (i.e. spatial collinearity) (Izpisúa-Belmonte et al., 1991). Interestingly, we noticed that the collinear sequence of Hoxb genes activation during gastrulation correlates with the temporal sequence of forelimb (e.g. Hoxb4) and interlimb (e.g. Hoxb7and Hoxb9) formation we had just identified, suggesting that they could play a role in the formation of these domains. To test it, we used the electroporation technique that allowed us to do functional perturbations precisely controlled in time and space. We performed overexpression and loss-of-function of different Hoxb genes in LPM precursor cells to perturb their activation during gastrulation. We could show that – as previously identified in the paraxial mesoderm (Iimura and Pourquié, 2006) – Hox genes, collinearly activated during gastrulation, establish their own stereotypical sequential expression domains in the LPM – domains that correlate with the forelimb (e.g. Hoxb4) and interlimb fields (e.g. Hoxb7 and Hoxb9).
But are these Hox expression domains important to position the forelimb?
One year before I started my PhD, a study showed that Hox4/5 genes could bind to a regulatory sequence of Tbx5 – a transcription factor essential for forelimb initiation – and activate the expression of a reporter gene under the control of this regulatory sequence (Minguillon et al., 2012). These results further supported a role for Hox genes in positioning the forelimb along the antero-posterior axis. We therefore decided to ectopically express Hoxb4 in the interlimb at stage 14 – when Hox domains are well-established – to see if we could perturb the forelimb position. We first screened embryos for an ectopic expression of Tbx5 in the interlimb, naively thinking that Hoxb4 overexpression should first induce Tbx5 expression. But that would have been too simple, Hoxb4 alone was not able to induce Tbx5 ectopic expression in the interlimb…
We were initially quite puzzled by this result. But the story could not stop there, we were missing something and we had to find out what. Then became a long journey during which we electroporated many different combinations of genes in the interlimb – e.g. Hoxb4 fused to VP16; combined Hox4/Hox5 genes; Hoxb4 plus activators of Retinoic Acid or Wnt/β catenin pathways, both involved in Tbx5 expression and forelimb initiation (Nishimoto et al., 2015); and several others – in an attempt to induce Tbx5 expression in the interlimb and a shift in limb position. Unfortunately, none of these combinations had an effect on Tbx5 expression nor limb position. At this point, we started to be somewhat desperate and were about to give up as the end of my PhD was getting closer. But reading again some of the literature, we came back to one paper in which it was shown that Hoxc9 – which is expressed in the interlimb – could repress Tbx5 expression (Nishimoto et al., 2014). Maybe the solution was there, this repressive activity was preventing all our attempts to posteriorly extend the Tbx5-positive forelimb field. This was it, the last-chance experiment! We decided to construct a dominant-negative form of Hoxc9 using a strategy others had established to generate Hox dominant-negative constructs (Denans et al., 2015). Then, we combined overexpression of Hoxb4 together with the repression of Hoxc9 in the interlimb and, not only we could nicely extend the Tbx5-positive forelimb domain in the interlimb, but we could displace the final forelimb position (Figure 1). There it was, many years after Hox genes where first suspected, we finally had the functional evidence of their direct role in regulating the forelimb position!
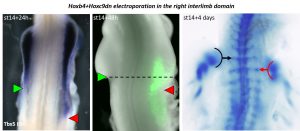
Importantly, removing Hoxc9 repression from the interlimb alone was not sufficient to induce Tbx5 expression and limb initiation. Therefore, showing that to change the forelimb position, both a shift of the forelimb field (e.g. Hoxb4 expression) and the interlimb field (e.g. Hox9 expression) is necessary. These results brought to us one potential explanation to why the vast majority of Hox genes mutant mice do not show major perturbations in limb position. Indeed, our data argue that, to induce a shift in the forelimb position in mouse, a combination of gain-of-function for forelimb activator (e.g. Hoxb4) and loss-of-function for forelimb-repressor (e.g. Hox9) should be performed. Another interesting point worth commenting is that, while Hoxb4 and Hox9 activate and repress Tbx5 expression, eventually leading to the establishment of the definitive forelimb position, Tbx5 itself was shown to be not sufficient to induce limb initiation (Nishimoto et al., 2015). Therefore, implying that, to regulate forelimb initiation at its specific position, Hox genes do not solely act upon activation/repression of Tbx5. This point remains to be explored but one can speculate that Hox genes could activate/repress other regulators of limb initiation or act cooperatively with Tbx5 to activate the limb initiation program, as recently shown for Hoxc10 and Tbx4 in the context of hindlimb development (Jain et al., 2018).
At this point, we had now unraveled the developmental mechanism behind limb position establishment. But what about limb position diversity, how can variations in limb position arise?
Our results suggested that natural variations in the forelimb position should be traced back to changes in Hox genes activation during gastrulation. To test this hypothesis, we took advantage of the bird natural variation in limb position. We selected three different bird species: zebra finch, chicken and ostrich as they form forelimbs at the level of the 13th, 15th and 18th vertebra, respectively, to perform a cross-species comparative analysis of Hoxb genes temporal and spatial dynamics of expression during early development. We then could provide evidence that, as predicted, changes in the timing of Hox activation during gastrulation prefigure variation in the spatial organization of these genes – i.e. in the spatial position of Hoxb4/Hoxb9 border of expression – in the LPM and ultimately, natural variation in forelimb position in birds. Working with non-conventional model organisms such as the ostrich was not trivial and required some troubleshooting and optimization at every step of the process, but it brought us with some very exciting outcomes and I think, shows the importance of using non-traditional model organisms when it comes to challenge and validate the models and predictions we establish using our favorite model organisms.
Finally, now that we had established the role of Hox genes in regulating limb position and variation, the next logical step was to investigate how such variation in Hox activation timing could be controlled. We got interested in the Retinoic Acid (RA) signaling pathway, and especially the RA catabolizing enzyme Cyp26a1, as it was already shown to be involved in hindlimb position regulation (Lee et al., 2010). We observed that Cyp26a1 onset of expression differs between the three avian species – i.e. its expression is premature and delayed in zebra finch and ostrich embryos, respectively, compared to chicken – consistent with a role for RA signaling in regulating Hox genes activation. Finally, modulating RA signaling during gastrulation in chicken embryos, provoked changes in Hox genes spatial organization in the LPM and in the Tbx5-positive forelimb field position, further suggesting that RA signaling might regulate the forelimb position through the regulation of Hox genes activation during gastrulation. This is an interesting point that will definitely need to be further investigated. Indeed, the possibility that Cyp26a1, already shown to regulate hindlimb position (Lee et al., 2010) could also regulate the forelimb position is particularly interesting. Especially, as it was recently proposed that differences in the onset of Gdf11 expression – i.e. the signaling molecule that induces Cyp26a1 expression – account for variations in hindlimb position amongst tetrapods (Matsubara et al., 2017). Therefore, one single signaling pathway would then be responsible for the regulation and natural variation of both fore- and hindlimb position.
Conclusion
From all these different results, the major conclusion of our work is that the forelimb position is determined very early, during gastrulation: it is the temporally controlled activation of Hox genes that progressively patterns the LPM into limb- and non-limb forming domains, as the main axis is being formed. And relative changes in this timed collinear activation underlie natural variation in forelimb position in birds (Figure 2).
One of the important points brought by our study is that we show the LPM is patterned by Hox genes during gastrulation following a similar mechanism as the one identified to pattern the somites (Iimura and Pourquié, 2006). These two tissues – that respectively give rise to the limb and vertebrae – both being generated and patterned during gastrulation by Hox genes, offers a simple mechanism to pattern the cervico-thoracic frontier in the somites and the forelimb position in the LPM, concomitantly therefore maintaining their tight association observed in all tetrapod species.
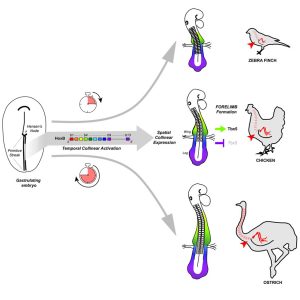
As a conclusion, our work that combines experimental embryology, state-of-the-art live imaging and cross-species comparative studies, addresses the major question of how the forelimb position is determined and solves a 20-year long controversy on the role of Hox genes in regulating limb position. It also provides a general mechanism for generating variation in body plan organization in vertebrates and reinforces the importance of Hox genes in shaping animal body plans.
Chloé Moreau
—————————
References
Burke, A.C., Nelson, C.E., Morgan, B.A., and Tabin, C. (1995). Hox genes and the evolution of vertebrate axial morphology. Dev. Camb. Engl. 121, 333–346.
Cohn, M.J., Patel, K., Krumlauf, R., Wilkinson, D.G., Clarke, J.D., and Tickle, C. (1997). Hox9 genes and vertebrate limb specification. Nature 387, 97–101.
Denans, N., Iimura, T., and Pourquié, O. (2015). Hox genes control vertebrate body elongation by collinear Wnt repression. ELife 4. e04379.
Iimura, T., and Pourquié, O. (2006). Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature 442, 568–571.
Izpisúa-Belmonte, J.C., Falkenstein, H., Dollé, P., Renucci, A., and Duboule, D. (1991). Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 10, 2279–2289.
Jain, D., Nemec, S., Luxey, M., Gauthier, Y., Bemmo, A., Balsalobre, A., and Drouin, J. (2018). Regulatory integration of Hox factor activity with T-box factors in limb development. Development 145, dev159830.
Lee, Y.J., McPherron, A., Choe, S., Sakai, Y., Chandraratna, R.A., Lee, S.-J., and Oh, S.P. (2010). Growth differentiation factor 11 signaling controls retinoic acid activity for axial vertebral development. Dev. Biol. 347, 195–203.
Mallo, M., Wellik, D.M., and Deschamps, J. (2010). Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344, 7–15.
Matsubara, Y., Hirasawa, T., Egawa, S., Hattori, A., Suganuma, T., Kohara, Y., Nagai, T., Tamura, K., Kuratani, S., Kuroiwa, A., et al. (2017). Anatomical integration of the sacral–hindlimb unit coordinated by GDF11 underlies variation in hindlimb positioning in tetrapods. Nat. Ecol. Evol. 1, 1392.
Minguillon, C., Nishimoto, S., Wood, S., Vendrell, E., Gibson-Brown, J.J., and Logan, M.P.O. (2012). Hox genes regulate the onset of Tbx5 expression in the forelimb. Dev. Camb. Engl. 139, 3180–3188.
Nishimoto, S., Minguillon, C., Wood, S., and Logan, M.P.O. (2014). A combination of activation and repression by a colinear Hox code controls forelimb-restricted expression of Tbx5 and reveals Hox protein specificity. PLoS Genet. 10, e1004245.
Nishimoto, S., Wilde, S.M., Wood, S., and Logan, M.P.O. (2015). RA Acts in a Coherent Feed-Forward Mechanism with Tbx5 to Control Limb Bud Induction and Initiation. Cell Rep. 12, 879–891.
Psychoyos, D., and Stern, C.D. (1996). Fates and migratory routes of primitive streak cells in the chick embryo. Development 122, 1523–1534.
Tanaka, M. (2013). Molecular and evolutionary basis of limb field specification and limb initiation. Dev. Growth Differ. 55, 149–163.
Tickle, C. (2015). How the embryo makes a limb: determination, polarity and identity. J. Anat. 227, 418-430.


 (2 votes)
(2 votes)