In Development this week (Vol. 141, Issue 6)
Posted by Seema Grewal, on 4 March 2014
Here are the highlights of the current issue of Development:
Auxin biosynthesis: the root of xylem patterning
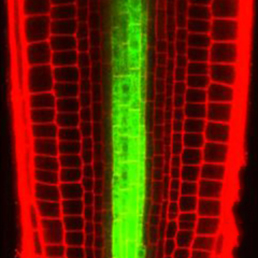 In the Arabidopsis root, xylem is organised into a central file of metaxylem that is flanked by protoxylem. Xylem fate is determined by HD-ZIP III transcription factors; high levels promote metaxylem formation whereas low levels specify protoxylem. The factors that downregulate HD-ZIP III levels are known but those that promote HD-ZIP III expression have remained elusive. Yunde Zhao, Ykä Helariutta, Jan Dettmer and colleagues now show that auxin biosynthesis promotes HD-ZIP III expression and metaxylem specification in Arabidopsis (p. 1250). The authors first isolate mutants that display defective xylem patterning and HD-ZIP III gene downregulation. These mutants harbour mutations in the gene encoding TRYPTOPHAN SYNTHASE BETA-SUBUNIT 1. Tryptophan is a precursor in the auxin biosynthesis pathway and, accordingly, the mutants exhibit aberrant auxin levels. They also show that metaxylem development and HD-ZIP III levels are defective in other auxin biosynthesis mutants. Based on their findings, the authors propose that tryptophan-dependent auxin biosynthesis is required for metaxylem formation.
In the Arabidopsis root, xylem is organised into a central file of metaxylem that is flanked by protoxylem. Xylem fate is determined by HD-ZIP III transcription factors; high levels promote metaxylem formation whereas low levels specify protoxylem. The factors that downregulate HD-ZIP III levels are known but those that promote HD-ZIP III expression have remained elusive. Yunde Zhao, Ykä Helariutta, Jan Dettmer and colleagues now show that auxin biosynthesis promotes HD-ZIP III expression and metaxylem specification in Arabidopsis (p. 1250). The authors first isolate mutants that display defective xylem patterning and HD-ZIP III gene downregulation. These mutants harbour mutations in the gene encoding TRYPTOPHAN SYNTHASE BETA-SUBUNIT 1. Tryptophan is a precursor in the auxin biosynthesis pathway and, accordingly, the mutants exhibit aberrant auxin levels. They also show that metaxylem development and HD-ZIP III levels are defective in other auxin biosynthesis mutants. Based on their findings, the authors propose that tryptophan-dependent auxin biosynthesis is required for metaxylem formation.
From Hippo to planarians
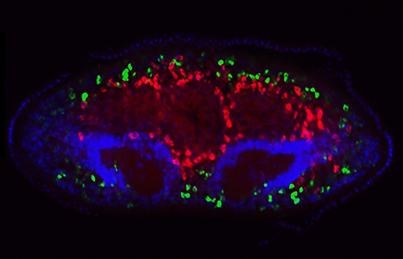 The transcriptional co-factor Yorkie (Yki; YAP in vertebrates) is a key effector of the Hippo signalling pathway and has been implicated in growth control and patterning, as well as stem cell regulation and regeneration, in flies and vertebrates. Now, on p. 1197, Alexander Lin and Bret Pearson investigate the role of Yki in planarian flatworms. The researchers report that planarians have a single orthologue of yki and, using RNAi, they show that yki carries out pleiotropic functions. For example, they report that ykiis required for homeostasis of the planarian excretory system, which is analogous to the vertebrate kidney. The researchers also show that, in contrast to its role in flies and vertebrates, yki functions to limit stem cell proliferation and hence the size of the stem cell population. In addition, yki plays a role in axial patterning, where it acts synergistically with Wnt signalling to supress head formation. These, together with other findings, demonstrate that yki plays diverse yet non-overlapping roles in planarian biology.
The transcriptional co-factor Yorkie (Yki; YAP in vertebrates) is a key effector of the Hippo signalling pathway and has been implicated in growth control and patterning, as well as stem cell regulation and regeneration, in flies and vertebrates. Now, on p. 1197, Alexander Lin and Bret Pearson investigate the role of Yki in planarian flatworms. The researchers report that planarians have a single orthologue of yki and, using RNAi, they show that yki carries out pleiotropic functions. For example, they report that ykiis required for homeostasis of the planarian excretory system, which is analogous to the vertebrate kidney. The researchers also show that, in contrast to its role in flies and vertebrates, yki functions to limit stem cell proliferation and hence the size of the stem cell population. In addition, yki plays a role in axial patterning, where it acts synergistically with Wnt signalling to supress head formation. These, together with other findings, demonstrate that yki plays diverse yet non-overlapping roles in planarian biology.
Following the leader
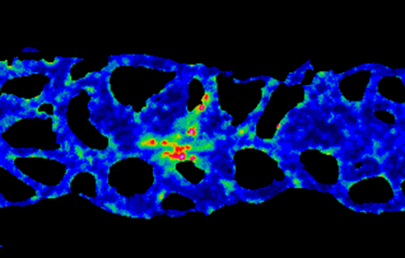 Collective cell migration occurs in many developmental contexts but a full understanding of this process has been hampered by a lack of quantitative analyses in 3D in vivo contexts. Using the zebrafish lateral line primordium as a model, Darren Gilmour and co-workers set out to tackle this problem (p. 1282). The researchers develop a method to simultaneously live-label microtubules, centrosomes and nuclei, allowing them to map cell polarity and orientation across the migrating population. Using this method, they identify the transition between leader cells and followers within the collective and show that this transition is marked by changes in cell-cell adhesion; cadherin 2 is expressed across the tissue but is only assembled into adherens junctions (AJs) in the transition zone and further down the leader-follower axis. A tandem fluorescent protein timer-based approach reveals that AJs become progressively more stable along the leader-follower axis. Finally, they show that AJ assembly, but not maintenance, requires dynamic microtubules, revealing a key role for microtubules during leader-to-follower transitions within migrating collectives.
Collective cell migration occurs in many developmental contexts but a full understanding of this process has been hampered by a lack of quantitative analyses in 3D in vivo contexts. Using the zebrafish lateral line primordium as a model, Darren Gilmour and co-workers set out to tackle this problem (p. 1282). The researchers develop a method to simultaneously live-label microtubules, centrosomes and nuclei, allowing them to map cell polarity and orientation across the migrating population. Using this method, they identify the transition between leader cells and followers within the collective and show that this transition is marked by changes in cell-cell adhesion; cadherin 2 is expressed across the tissue but is only assembled into adherens junctions (AJs) in the transition zone and further down the leader-follower axis. A tandem fluorescent protein timer-based approach reveals that AJs become progressively more stable along the leader-follower axis. Finally, they show that AJ assembly, but not maintenance, requires dynamic microtubules, revealing a key role for microtubules during leader-to-follower transitions within migrating collectives.
Dbx1 crosses the (mid)line
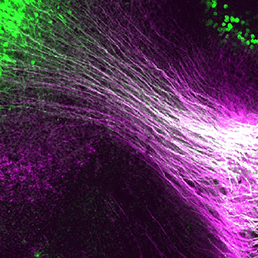 During nervous system development, navigating axons ‘decide’ whether or not to cross the midline. Various factors that influence axon guidance and midline crossing have been identified but it remains unclear if any one transcription factor can drive the complete midline crossing transcriptional programme. Here (p. 1260), Yasuyuki Inamata and Ryuichi Shirasaki report that a single homeodomain transcription factor, Dbx1, assigns midline-crossing identity at the progenitor stage in mice. The researchers show that Dbx1 is expressed in a subset of neural progenitors in the dorsal midbrain. Lineage tracing demonstrates that midbrain commissural neurons, which cross the midline, are generated selectively from Dbx1-positive progenitors. Furthermore, gain- and loss-of-function experiments show that Dbx1 is necessary and sufficient for midline crossing. Finally, the authors show that Dbx1 controls a molecular programme that controls the expression of Robo3, an essential regulator of midline crossing, on commissural neurons while repressing the ipsilateral neuron genetic programme. These findings reveal an unanticipated regulatory layer within the transcriptional cascade that controls nervous system wiring.
During nervous system development, navigating axons ‘decide’ whether or not to cross the midline. Various factors that influence axon guidance and midline crossing have been identified but it remains unclear if any one transcription factor can drive the complete midline crossing transcriptional programme. Here (p. 1260), Yasuyuki Inamata and Ryuichi Shirasaki report that a single homeodomain transcription factor, Dbx1, assigns midline-crossing identity at the progenitor stage in mice. The researchers show that Dbx1 is expressed in a subset of neural progenitors in the dorsal midbrain. Lineage tracing demonstrates that midbrain commissural neurons, which cross the midline, are generated selectively from Dbx1-positive progenitors. Furthermore, gain- and loss-of-function experiments show that Dbx1 is necessary and sufficient for midline crossing. Finally, the authors show that Dbx1 controls a molecular programme that controls the expression of Robo3, an essential regulator of midline crossing, on commissural neurons while repressing the ipsilateral neuron genetic programme. These findings reveal an unanticipated regulatory layer within the transcriptional cascade that controls nervous system wiring.
Lymphangiogenesis: of mice and fish
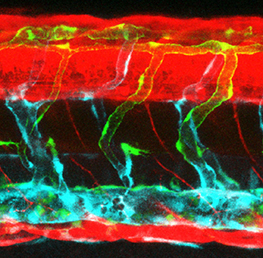 In mice, the formation of lymphatic vessels (lymphangiogenesis) requires the homeodomain transcription factor Prox1. Here, Stefan Schulte-Merker and colleagues examine whether the role of Prox1 is conserved in zebrafish (p. 1228). Using a novel transgenic reporter line, the researchers show that, in contrast to the situation seen in mice, zebrafish Prox1 is initially not expressed in all lymphatic precursor cells and reliably marks this population only during later stages of lymphangiogenesis, arguing against a role for Prox1 in lymphatic specification. In addition, targeted mutagenesis demonstrates that lymphangiogenesis can proceed in the complete absence of Prox1. Finally, they show that the functionally related transcription factors Coup-TFII and Sox18, which are implicated in lymphatic specification in mice, are also dispensable for zebrafish lymphangiogenesis. The authors conclude that an alternative lymphatic specification mechanism is present in zebrafish and propose that differences in the timing of lymphangiogenesis between mice and fish can explain this divergence.
In mice, the formation of lymphatic vessels (lymphangiogenesis) requires the homeodomain transcription factor Prox1. Here, Stefan Schulte-Merker and colleagues examine whether the role of Prox1 is conserved in zebrafish (p. 1228). Using a novel transgenic reporter line, the researchers show that, in contrast to the situation seen in mice, zebrafish Prox1 is initially not expressed in all lymphatic precursor cells and reliably marks this population only during later stages of lymphangiogenesis, arguing against a role for Prox1 in lymphatic specification. In addition, targeted mutagenesis demonstrates that lymphangiogenesis can proceed in the complete absence of Prox1. Finally, they show that the functionally related transcription factors Coup-TFII and Sox18, which are implicated in lymphatic specification in mice, are also dispensable for zebrafish lymphangiogenesis. The authors conclude that an alternative lymphatic specification mechanism is present in zebrafish and propose that differences in the timing of lymphangiogenesis between mice and fish can explain this divergence.
Fresh air in the mesenchyme
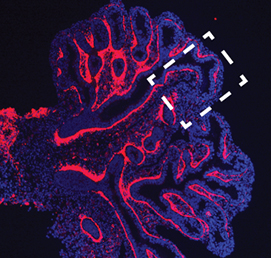 The mesenchymal compartment of the lung plays a crucial role during lung development but, unlike its epithelial counterpart, its regulation is largely uncharacterised. Now, Gianni Carraro, Saverio Bellusci and colleagues report that miR-142-3p modulates WNT signalling to balance mesenchymal cell proliferation and differentiation during mouse lung development (p. 1272). Using microarray analyses, the researchers identify miR-142-3p as highly expressed in the embryonic lung mesenchyme. Importantly, loss-of-function assays demonstrate that miR-142-3pregulates cell proliferation specifically in the mesenchyme; in the absence of miR-142-3p, progenitor cells prematurely differentiate. They also show that miR-142-3p binds to and regulates the expression of mRNA encoding APC, a negative regulator of WNT signalling. Accordingly, miR-142-3p knockdown can be rescued by activating WNT or reducing APC expression in the mesenchyme. Based on their findings, the authors propose that miR-142-3p adds an extra layer of control to the WNT-FGF feedback loop that operates in the lung mesenchyme to correctly balance cell proliferation and differentiation.
The mesenchymal compartment of the lung plays a crucial role during lung development but, unlike its epithelial counterpart, its regulation is largely uncharacterised. Now, Gianni Carraro, Saverio Bellusci and colleagues report that miR-142-3p modulates WNT signalling to balance mesenchymal cell proliferation and differentiation during mouse lung development (p. 1272). Using microarray analyses, the researchers identify miR-142-3p as highly expressed in the embryonic lung mesenchyme. Importantly, loss-of-function assays demonstrate that miR-142-3pregulates cell proliferation specifically in the mesenchyme; in the absence of miR-142-3p, progenitor cells prematurely differentiate. They also show that miR-142-3p binds to and regulates the expression of mRNA encoding APC, a negative regulator of WNT signalling. Accordingly, miR-142-3p knockdown can be rescued by activating WNT or reducing APC expression in the mesenchyme. Based on their findings, the authors propose that miR-142-3p adds an extra layer of control to the WNT-FGF feedback loop that operates in the lung mesenchyme to correctly balance cell proliferation and differentiation.
PLUS…
Growing older gracefully – a review of the 10th edition of Developmental Biology
 First published in 1985, Developmental Biology by Scott F. Gilbert has been an influential textbook for a generation of scientists. Here, Timothy Weil provides a brief review of the latest (10th) edition of the series. He comments on updates to the text, highlights details of particular interest to lecturers, and compares this book with other resources available in the internet era. See the Spotlight article on p. 1177
First published in 1985, Developmental Biology by Scott F. Gilbert has been an influential textbook for a generation of scientists. Here, Timothy Weil provides a brief review of the latest (10th) edition of the series. He comments on updates to the text, highlights details of particular interest to lecturers, and compares this book with other resources available in the internet era. See the Spotlight article on p. 1177
RNA polymerase II pausing during development
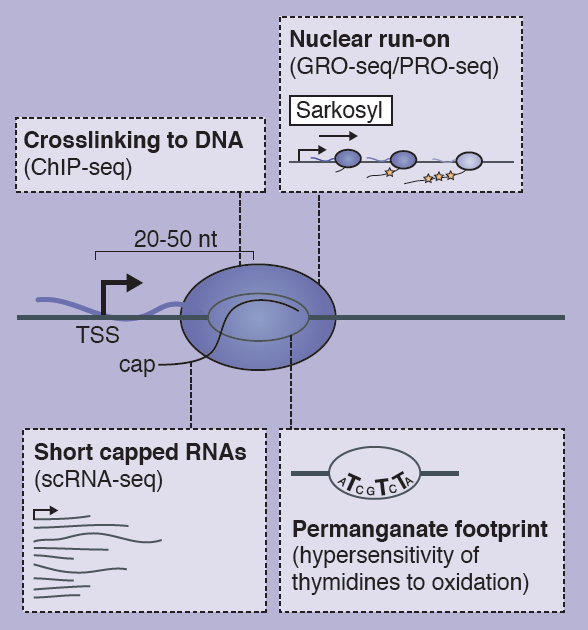 The transcription of developmental control genes by RNA polymerase II (Pol II) is commonly regulated at the transition to productive elongation, resulting in the promoter-proximal accumulation of transcriptionally engaged but paused Pol II. In their poster article, Bjoern Gaertner and Julia Zeitlinger review the mechanisms and possible functions of Pol II pausing during development. See the Development at a Glance article on p. 1179
The transcription of developmental control genes by RNA polymerase II (Pol II) is commonly regulated at the transition to productive elongation, resulting in the promoter-proximal accumulation of transcriptionally engaged but paused Pol II. In their poster article, Bjoern Gaertner and Julia Zeitlinger review the mechanisms and possible functions of Pol II pausing during development. See the Development at a Glance article on p. 1179
Shared signaling systems in myeloid cell-mediated muscle regeneration
![]() Much of the focus in muscle regeneration has been placed on identifying and delivering stem cells to promote regenerative capacity. As these efforts have advanced, we have learned that complex features of the microenvironment in which regeneration occurs can determine the success of these approaches . Here, James Tidball and colleagues discuss how myeloid cells can influence muscle regeneration, focussing on how processes in muscle and myeloid cells are co-regulated. See the Review on p. 1184
Much of the focus in muscle regeneration has been placed on identifying and delivering stem cells to promote regenerative capacity. As these efforts have advanced, we have learned that complex features of the microenvironment in which regeneration occurs can determine the success of these approaches . Here, James Tidball and colleagues discuss how myeloid cells can influence muscle regeneration, focussing on how processes in muscle and myeloid cells are co-regulated. See the Review on p. 1184


 (1 votes)
(1 votes)