An interview with Bill Harris
Posted by the Node, on 6 July 2017
This interview by Aidan Maartens originally appeared in Development, Volume 143, Issue 13.
William ‘Bill’ Harris is Head of the Department of Physiology, Development and Neuroscience at the University of Cambridge, UK, and a Fellow of both the Royal Society and Academy of Medical Sciences. His lab works on the development of the vertebrate nervous system, with a particular focus on cell lineage in the retina. In 2017 he was awarded the British Society for Developmental Biology’s Waddington Medal for outstanding research performance and services to the community. We met Bill in his Cambridge lab to talk science, art and ice hockey.

This year you were awarded the Waddington Medal: what does the award mean to you?
Well, it was a signal that developmental biologists appreciated what I’ve done over my career. I’ve always considered myself first as a neuroscientist, but a developmental neuroscientist, and as I’ve progressed in my career I’ve come to appreciate that the problems I’ve been working on in neuroscience are problems that are common to lots of developmental systems. So to get the appreciation of the field as a whole is very gratifying. It’s the same kind of feeling you get whenever something you’ve done gets accepted, whether a paper or a grant or whatever, that what you’ve done has got through certain hurdles and been deemed OK. Also, as it was near the end of my career and was basically a lifetime achievement award, I was particularly pleased about that.
Let’s go back to the beginning: what got you interested in science, and neuroscience in particular, in the first place?
I got interested in science when I was an undergraduate at the University of California Berkeley, though I guess you could say that before that I was as interested in science as any other kid might be. I did a senior project using one of the early scanning electron microscopes, and decided to look at Drosophila, knowing that no one had ever seen fruit flies blown up to this magnification before. All of a sudden I could see these hidden things, and I knew that geneticists would be interested in them – it was a bit of real, useful science, but was also just amazing to see that level of detail in such a small organism. I keep an image of a Drosophila head from that project up on my office wall to this day.
So that project got me interested in science, but getting interested in the development of the nervous system was a completely different ball game – I got there by a more osmotic, slow process. As a PhD student with Seymour Benzer at CalTech, I was interested in behaviour and genes, in the function of the nervous system. I worked on learning mechanisms in flies, and spent a lot of time in a little dark room getting flies to run to different coloured lights and punishing them if they went to some lights and not others. Then I got some advice that I would spend my whole graduate career just learning about the psychology of flies and not learning any biology if I kept on with that project, so I switched to looking at the fly visual system, and found some mutants that didn’t see certain colours. That got me into the cells, the molecules, the photochemistry and physiology – it was a door into ‘wet’ biology and into the anatomy of the nervous system. That was a better fit for me – I felt I was really learning something.

How did you come to work on the vertebrate nervous system?
As a postdoc I went to Harvard Medical School to work in the labs of David Hubel and Torsten Weisel, who were doing beautiful work on the way the visual cortex operates and the relationship between the physiology of the cortex and its cytoarchitecture. I got interested in the developmental problem of how the early function of the nervous system shapes the way it continues to develop. It was a very exciting time, and the postgrads and postdocs were doing all kinds of interesting, fun experiments. But then one day we were called into the tea room and got a lecture from Hubel and Weisel, who said we shouldn’t just do all these different projects – some of which we had started without even consulting them – and that we were just using their labs as a launching point for our things, whereas they had their own vision for where they wanted the lab to go. They said ‘We’ve planted this plum tree, and now all the plums are very ripe and you guys are just picking them off without even asking!’. Although some of the people were absolutely committed to the visual cortex and didn’t know why they were being told off for doing interesting experiments, it changed my approach to my work – I understood somehow that I should be more independent and that I should try to find my own niche.

So that’s when I switched to a more developmental programme of interests. I moved to axolotls first, largely due to Of Scientists and Salamanders, a book by Victor Twitty, which showed me the wonders of salamander developmental biology. Twitty had found a species of Californian newt that was full of tetrodotoxin, and if you parabiotically joined an embryo of that species to the embryo of an axolotl, the toxin would percolate from the newt embryo (which was insensitive to the toxin) to the axolotl embryo, blocking all of the neural activity in the latter. That allowed me to get back to the earlier question of the role of neural activity in development, which led me to do an experiment revealing that a lot of neural development, including accurate wiring up of the nervous system, did not require any electrical activity. This was an important step forward for the field, which previously thought that neural activity was key to making appropriate connections in the brain. I then started to work on axon guidance – since axons can get to their targets in the absence of neural activity, how exactly do they get there?
So I started with behaviour and have kept going backwards in time until where I am now, which is to ask how different cells in the nervous system get born. That’s probably as far back as I’m going to go or else I’ll move out of the nervous system altogether!
I started with behaviour and have kept going backwards in time until where I am now, which is to ask how different cells in the nervous system get born
Since the early eighties, your lab has mainly focussed on the development of the vertebrate retina. With your wife Christine Holt, you described techniques for labelling neurons to determine lineage and fate – what were the main lessons this early labelling taught you?
In 1980 I started as an Assistant Professor at the University of California San Diego, and a bit later on Christine came to the lab as a postdoc, bringing Xenopuswith her. I used to have a nice life before Christine, in that for the axolotl work you’d get the embryos in the autumn, do your embryonic manipulations, wait for them to grow until you could see what had happened later in the year, then work on writing up these results until the next autumn when the cycle started again. With Xenopus, Christine found a way that you could label specific populations of axons even when they were growing, and not only that, but you could get embryos all year round in the lab – now you could do experiments any day you wanted to and get the results as early as the next day! It was a much more powerful system and allowed the work to go at a faster pace, making the work more exciting and more demanding.
We were interested in the very earliest decisions that retinal ganglion cells made as they were navigating the pathway into the brain. We thought about injecting the cells to label them at earlier and earlier stages, just after they were born, so you could see the first thing the cell did after it was born and knew it was going to be a retinal ganglion cell, to ask how its axon started the journey. We then realised we could inject the cell even before it was born, to look at the mother cell. At the same time, Connie Cepko’s lab had started to use replication-incompetent retroviruses to look at similar problems, and Scott Fraser’s lab had similar ideas, also in Xenopus. We all ended up publishing at around the same time, and had more or less the same results.
The main lesson this labelling taught us was that progenitors in the retina and in the brain were multipotent – they would give rise to many different cell types, ruling out the possibility that there were committed progenitors for each cell type. One thing that puzzled all of us, and led us in the wrong direction for quite a while, was the variability of the lineages – you’d never get the same lineage twice. We all reported this, but didn’t understand it – it might have had to do with different cells experiencing different microenvironments, but it turns out it is a much more intrinsic thing.
In recent years you have had a productive collaboration with Benjamin Simons from the Cavendish Laboratory – how important has mathematical modelling been to your understanding of cell fate in the retina?
It’s shaped the way I think about the variable nature of cell lineages in the retina. Ben came to me after working with Phil Jones on the epidermis of the mouse tail: he was looking at tissues that are homeostatic, where you are losing cells and replacing them at the same rate. Being a physicist he could look at the cell fate data and somehow see in these patterns the underlying model. So he came to me and said that the retina could be a great model to understand how lineages evolve, but I thought that this would probably be too difficult because, during development, things are moving and the rules are going to be changing all the way through. Jie Hie (a postdoc in the lab) and I listened very carefully to the kind of data that he would need, which he could analyse powerfully, and figured out how we might be able to provide it. The analysis revealed that there is very likely a stochastic phase in the development of clones of retinal cells which produces the variability – now we have a model whereby a period of stochasticity means that genes may or may not get expressed, and that this then affects fate in a probabilistic way.
The underlying mathematics isn’t really that hard, but to actually run the simulations and statistically compare the results against the real data is something that Ben is more of an expert at than I. Back when we first published our early clonal data and saw this variability, all I was really able to say was that you can’t discriminate our data from just randomising the numbers of cells that went into the different layers; Ben’s analysis gave us a model that suggests something about the biology. So it really helped to understand those things, and is still helping to motivate work that we’re doing.
In your acceptance speech for the Waddington Medal, you compared the models for cell fate that came out of your work with Waddington’s famous epigenetic landscape. How do Waddington’s ideas relate to your current understanding of stochasticity and determinism in retinal development?
Every developmental biology student has seen Waddington’s epigenetic landscape drawing – it’s a wonderful way to think about development in a diagrammatic way.
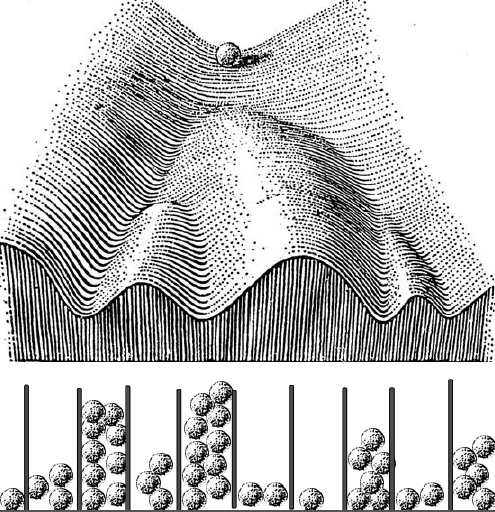
As I was preparing the lecture I wondered what Waddington thought about how cells made decisions to go down one valley or another, but I could not really find much – he described the geography of cell fate decisions, but not how cells made the decisions. His one vague suggestion was that something might be pushing the marbles toward one valley or another. But I was thinking about these probabilities that we’ve been working on, and asked myself if there was a way to join his landscape with probability distributions. As I was thinking about this, my mind went to these Galton boards – you drop a marble, it hits a pin and goes to the right or the left, and then hits another pin and does the same thing, and at the end of that, if you put enough marbles in, you get a beautiful normal distribution in the wells at the bottom. It’s a great way of creating a distribution that is always the same shape even though any particular marble could end up in any of the wells – and if you want to make an organism that’s the same, you want to make a distribution of cell types that’s the same. So let’s run these marbles down through the Waddington landscape, which is really a kind of a Galton board – at each junction it has two choices – but the probabilities are set because of the deepness of the valley and how the ball was rolling. The important thing is that you won’t get the same balls in the same valleys in every organism, but you’ll get the same distribution. So in the talk I just took Waddington’s epigenetic landscape and turned it into a Galton board.
The Waddington Medal lecture can be viewed here
You have been Head of the Department of Physiology, Development and Neuroscience in Cambridge since its inception in 2006. What has it been like steering such a diverse department of researchers?
I came to Cambridge in 1997 as Professor of Anatomy, and the next year I started to take over the reins as Head of Department. In 2006, Anatomy merged with Physiology; there were a lot of similarities in terms of the students we taught and the kinds of research that went on in both departments, so it was a natural union. I think the merger gave a new lease of life to the two departments, and it also helped us with scales of efficiency as we were both running in the red at the time. I don’t consider it a particular challenge to be Head of Department – it’s quite easy if people are motivated to help make the department better. It’s important that everyone has a say in how the place is run – so in meetings I prefer to present ideas and see what everyone thinks, and then together we’ll agree on a way forward democratically.
How did the Cambridge Advanced Imaging Centre (CAIC) come about?
When I came here there was already an installation downstairs that had two electron microscopes, and at the time I was becoming increasingly interested in issues of lineage and taken with making movies of development. We initially applied for a two-photon microscope as a tool to visualise these things. There were clearly advances in light microscopy that were going on and it was important to me that there was a facility in Cambridge that offered top-end microscopy for biologists. I also knew that there were people in the physical sciences at Cambridge that were really interested in advancing microscopy, without necessarily being interested in biological questions. CAIC was an idea that came out of these interactions – we wanted to build a new generation of advanced light microscope instruments specifically for biologists, and quickly tuneable to their specific needs. So in CAIC we build microscopes not just to see how far microscopy can go, but to tackle crucial issues in biology. It has become an amazing facility that a lot of people are using, and is a hub of a wider set of microscopy centres around the university.
You enjoy painting, and in studying the visual system your lab generates beautiful images of retinal organisation. Have you always been a visually oriented person, and has this directed your research?
Since I was a kid I’ve always liked to draw, though I wouldn’t consider myself a particularly talented artist. The science that I’ve done has always depended on imaging – from that very first time when I looked through the scanning electron microscope as an undergraduate, I was hooked on seeing the microscopic world. I can appreciate bands on gels and things like that, but I like to see the cells, and the questions that I’ve ended up asking over the years – like how do axons get from A to B, or how lineages arise – mean that I want to see the processes occurring. I like to see things in three or four dimensions, and the prettier they are the better it is somehow. I particularly love it when a student or postdoc runs into the office saying ‘Bill, you’ve gotta see this!’, and takes me over to the microscope to show me what’s going on.

In my painting – which I turned to again about ten years ago as a serious hobby, in preparation for my retirement – I took some of my doodles, bits of retina and things like that, and started to colour them in with paints. I think I will continue to do biology-themed paintings for a while now.
From that very first time when I looked through the scanning electron microscope as an undergraduate, I was hooked on seeing the microscopic world
I hear you’re into ice hockey? It must be quite a niche pursuit in Cambridge?
In fact, there are quite a few people playing ice hockey in the UK: it’s a niche sport but it’s growing. I grew up as a Canadian, and was on skates when I was four or five – we used to flood our back yard to make an ice rink, and I would play ice hockey with my brothers. I was never a great player, like I’m not a great painter, but I liked it enough that I kept doing it. When I came here to Cambridge, I discovered there was a team – in fact, the oldest ice hockey rivalry in the world is between Oxford and Cambridge. So I quickly got involved, became the coach of the team, and also play in the recreational team, though there I’m more of a hazard than a help.

Is there anything that Development readers would be surprised to find out about you?
I am the youngest of four children – an older sister and two older brothers. So I was the baby of the family and have always been treated that way. I keep thinking that after all my years of experience as Head of Department, and in life more generally, I’m going to meet them one day and be able to hold my own. But no, they still treat me as a useless baby brother who doesn’t know anything and is not worth listening to. After all these years, it hasn’t changed!


 (2 votes)
(2 votes)