Fight for Sight PhD position: Identification of novel genes involved in fevea formation-London, UK
Posted by Florencia Cavodeassi, on 6 May 2024
Location: London, UK
Closing Date: 26 May 2024
Start date: 1 October 2024
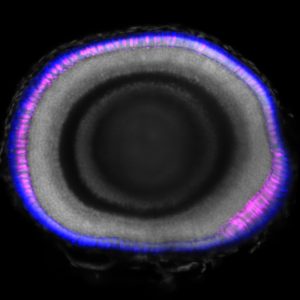
Florencia Cavodeassi (St George’s, University of London) and Gaia Gestri (University College London) have been awarded a 4-year Fight for Sight PhD Studentship with the title “Identification of novel genes involved in fovea formation”, to start on the 1st October 2024. The project follows up from the research article “Foxd1-dependent induction of a temporal retinal character is required for visual function”, published in Development in December 2022.
Project details
Visual acuity depends on the fovea, located at the back of the retina, in the centre of a structure called the macula. Congenital ocular diseases resulting from genetic mutations that affect macula formation during embryogenesis can disrupt the fovea and lead to a defect called foveal hypoplasia, associated to visual deficiency.
Most cases of congenital foveal hypoplasia lack a genetic diagnosis, limiting our ability to predict disease progression or provide patient counselling. Improving genetic diagnosis requires identifying those genes important for fovea formation, and uncovering how they work. We use the zebrafish to tackle these questions. Zebrafish, like humans, bear in their retina an area specialised in high acuity vision. In previous work, we generated a genetically modified strain of zebrafish with a mutation in a gene involved in the formation of the high acuity area. By comparing the genes that are turned on in the retina of these mutants with those genes turned on in wild type zebrafish, we have identified a subset of genes that may be important for the formation and function of the zebrafish high acuity area.
The aim of the project is to advance our understanding of the mechanisms controlling fovea formation by characterising the molecular and behavioural function of those novel genes. In addition, together with our clinical collaborators, we will search in their cohorts of patients with ocular disease for potential variants in our genes of interest. We will also work with computational geneticists to search databases such as the 100K Genome Project and GeneMatcher, to identify further individuals with potentially pathogenic variants in our genes of interest.
Skills acquisition
The student will be primarily based at St George’s, but experiments will also be performed at UCL, where one of the co-supervisors is based. The student will be trained in the latest approaches in gene editing and mis-expression to abrogate gene function in zebrafish. They will then analyse the effect of these manipulations with molecular and imaging approaches. In addition, visual function will be assessed by subjecting the larvae to visual performance tests.
The student will be trained in robust computational analysis. Together with our clinical collaborators, they will search in their cohorts of patients with ocular disease for potential variants in the genes of interest. They will also search databases such as the 100K Genome Project and GeneMatcher, to identify further individuals with potentially pathogenic variants in our genes of interest.
For more information on funding details and application process follow this link: https://www.sgul.ac.uk/study/postgraduate-study/research-degrees/phd-projects/fight-for-sight-phd-studentship-2024-25
Start date: 1 October 2024
Closing Date: 26 May 2024
Scientific fields: Development and disease, Organogenesis, Patterning
Model systems: Zebrafish
Duration: Fixed term

