PhD position “Imaging long-distance communication between homologous alleles to unravel sex-specific mechanisms of gene regulation” – Prudhomme Lab, IBDM, Marseille, France
Posted by Delphine Dauga, on 8 June 2021
Location: IBDM, Marseille, France
Closing Date: 30 September 2021
The Prud’homme team is welcoming application for one PhD student position to work on a collaborative imaging and modeling project with the lab of Thomas Gregor (Institut Pasteur, Princeton University).
Research Background:
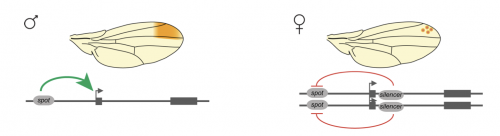
The expression of a gene is regulated by its flanking cis-regulatory sequences. However, proper gene expression relies also often on interallelic gene regulation. How alleles sitting on homologous chromosomes find each other in the nucleus, and how their interactions influence gene expression are open questions. We study these general questions by focusing on the regulation of the Drosophila gene yellow [1]. yellow is located on the X chromosome, allowing the two alleles to interact in XX females but not on XY males. Thus, female-specific interallelic interactions shape the sex-specific expression of yellow [2].
Research objectives:
The proposed project aims at visualizing the interaction between homologous yellow alleles and the functional consequences of that interaction on gene expression. Super-resolution imaging in fixed cells will elucidate spatial conformations of the DNA polymer and associate these to gene regulatory states; in live cells the goal is to assess the temporal dynamics of gene expression and how it is linked to fluctuating distances between the two homologous polymer strands. For more information about these approaches see [3, 4].
Research environment:
This interdisciplinary project will be carried out in close collaboration between the Prud’homme lab at IBDM, in Marseille, and the Gregor lab at Institut Pasteur, in Paris. These two dynamic places are internationally-recognized for their interdisciplinary research environments and offer excellent training opportunities for students.
Your profile:
The project is best suited for a self-driven and independent student, seeking a highly interdisciplinary education; either with a background in physics and a strong interest for biological questions, or with a biology education with a strong background in imaging and computational analysis. Coding skills and statistical training are strongly recommended. The position has to be filled by Dec. 1st 2021.
To apply for this position, please send a CV, a letter of motivation, and contacts for references to:
More information about each group can be found here:
References:
[1] Arnoult, L., et al. (2013). Science 339, 1423–1426. doi: 10.1126/science.1233749
[2] Galouzis, C.C., and Prud’homme, B. (2021). Science 371, 396–400. doi.org/10.1101/2020.03.23.003103 [3] Chen, H., et ak. (2018). Nature Gene6cs 50, 1296–1303. doi: 10.1038/s41588-018-0175-z
[4] Barinov, L., et al. (2020). arXiv:2012.15819 [q-bio].
Annoucement in pdf
Closing Date: 30 September 2021
Scientific fields: Gene regulation
Model systems: Drosophila
Duration: Fixed term

