March in preprints
Posted by the Node, on 4 April 2018
One of the most talked about preprints this month was a corrigendum to that Nature Methods paper reporting widespread off-target mutations following CRISPR-Cas9 editing in mice. The paper was challenged quickly on bioRxiv, and four days after the corrigendum went up, it was retracted, as reported by Retraction Watch! Blink and you’ll miss it.
Otherwise this month we found plenty of plant patterning, lovely larval lampreys and ovulating roundworms, and the developing diaphragm finding its voice.
The preprints were hosted on bioRxiv, PeerJ, and arXiv. Use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice & education
Why not…
Developmental biology
| Patterning & signalling
Positional information encoded in the dynamic differences between neighbouring oscillators during vertebrate segmentation.
Marcelo Boareto, Tomas Tomka, Dagmar Iber
Checkpoint signaling abrogation after cell cycle reentry reveals that differentiated neurons are mitotic cells
Chaska Carlos Walton, Wei Zhang, Iris Patiño-Parrado, Estíbaliz Barrio-Alonso, Juan-José Garrido, José María Frade
Epidermal YAP Activity Drives Canonical WNT16/β-catenin Signaling to Promote Keratinocyte Proliferation in vitro and in the Murine Skin
Veronica Mendoza-Reinoso, Annemiek Beverdam
Extracellular Pax6 regulates tangential Cajal-Retzius cell migration in the developing mouse neocortex
Hadhemi Kaddour, Eva Coppola, Ariel Di Nardo, Andrea Wizenmann, Michel Volovitch, Alain Prochiantz, Alessandra Pierani
Megan B O’Hare, Alamin Mohammed, Kyle J Connolly, Katelyn C Aitchison, Niki C Anthoney, Amy L Roberts, Matthew J Taylor, Bryan A Stewart, Richard I Tuxworth, Guy Tear
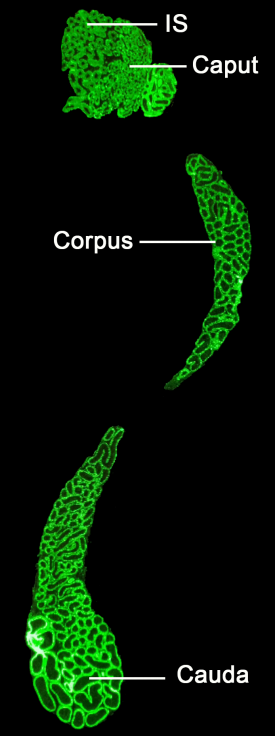
Cell-lineage specificity of primary cilia during epididymis post-natal development
Agathe Bernet, Alexandre Bastien, Denis Soulet, Olivia Jerczynski, Christian Roy, Maira Bianchi Rodrigues Alves, Cynthia Lecours, Marie-Eve Tremblay, Janice Bailey, Claude Robert, Clemence Belleannee
Hideki Katow, Hiromi Yoshida, Tomoko Katow, Masato Kiyomoto
Embryonic and postnatal neurogenesis produce functionally distinct subclasses of dopaminergic neuron
Elisa Galliano, Eleonora Franzoni, Marine Breton, Annisa N. Chand, Darren J. Byrne, Venkatesh N. Murthy, Matthew S. Grubb
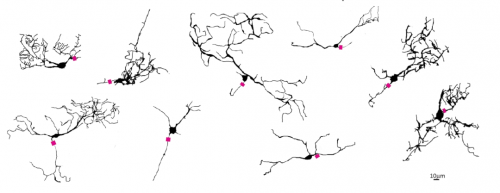
Formation of retinal direction-selective circuitry initiated by starburst amacrine cell homotypic contact
Thomas A Ray, Suva Roy, Christopher Kozlowski, Jingjing Wang, Jon Cafaro, Samuel Hulbert, Christopher V E Wright, Greg D Field, Jeremy N Kay

Conservation and Divergence of YODA MAPKKK Function in Regulation of Grass Epidermal Patterning
Emily Abrash, Maria Ximena Anleu Gil, Juliana L Matos, Dominique C Bergmann
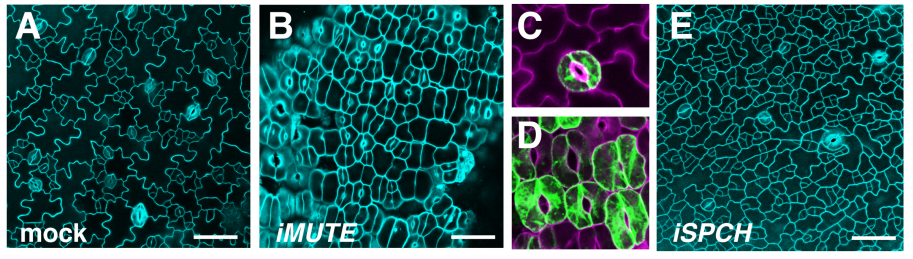
MUTE Directly Orchestrates Cell State Switch and the Single Symmetric Division to Create Stomata
Soon-Ki Han, Xingyun Qi, Kei Sugihara, Jonathan Dang, Takaho A Endo, Kristen Miller, Eun-Deok Kim, Takashi Miura, Keiko Torii
Clonal analysis reveals gradual recruitment of lateral root founder cells and a link between root initiation and cambium formation in Arabidopsis thaliana
Joseph G. Dubrovsky
Auxin is not asymmetrically distributed in initiating Arabidopsis leaves
Neha Bhatia, Marcus G Heisler
A Lotus japonicus E3 ligase interacts with the Nod factor receptor 5 and positively regulates nodulation
Daniela Tsikou, Estrella E. Ramirez, Ioanna S Psarrakou, Jaslyn E Wong, Dorthe B Jensen, Erika Isono, Simona Radutoiu, Kalliope K Papadopoulou
Pollen Patterns Form from Modulated Phases
Asja Radja, Eric M. Horsley, Maxim O. Lavrentovich, Alison M. Sweeney
A nuclear hormone receptor and lipid metabolism axis are required for the maintenance and regeneration of reproductive organs
Shasha Zhang, Longhua Guo, Carlos Guerrero-Hernández, Eric J Ross, Kirsten Gotting, Sean A. McKinney, Wei Wang, Youbin Xiang, R. Scott Hawley, Alejandro Sánchez Alvarado
PIP5k1 β controls bone homeostasis through modulating both osteoclast and osteoblast differentiation
Xiao-ying Zhao, Guoli Hu, Chuandong Wang, Lei Jiang, Jingyu Zhao, Jiake Xu, Xiaoling Zhang
Analysis of novel domain-specific mutations in the zebrafish ndr2/cyclops gene generated using CRISPR-Cas9 RNPs
Ashley N Turner, Reagan S Andersen, Ivy E Bookout, Lauren N Brashear, James C Davis, David M Gahan, John P Gotham, Baraa A Hijaz, Ashish S Kaushik, Jordan B McGill, Victoria L Miller, Zachariah P Moseley, Cerissa L Nowell, Riddhi K Patel, Mia C Rodgers, Yazen A Shihab, Austin P Walker, Sarah R Glover, Samantha D Foster, Anil Kumar Challa
Deep proteomic analysis of chicken erythropoiesis
Marjorie Leduc, Emilie-Fleur Gautier, Anissa Guillemin, Cédric Broussard, Virginie Salnot, Catherine Lacombe, Olivier Gandrillon, François Guillonneau, Patrick Mayeux
PRY-1/AXIN signaling regulates lipid metabolism in Caenorhabditis elegans
Ayush Ranawade, Avijit Mallick, Bhagwati P Gupta
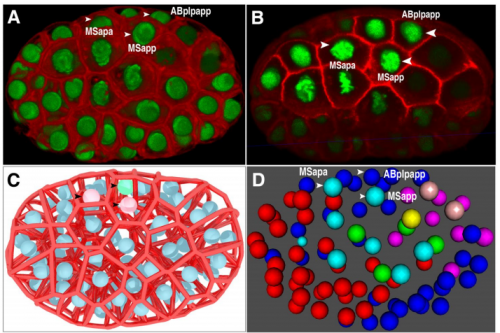
Establishment of signaling interactions with cellular resolution for every cell cycle of embryogenesis
Zhongying Zhao, Long Chen, Vincy WS Ho, Ming-Kin Wong, Xiaotai Huang, Lu-yan Chan, Kaoru HC Ng, Xiaoliang Ren, Hong Yan
Murine SEC24D Can Substitute Functionally for SEC24C in vivo
Elizabeth J. J. Adams, Rami Khoriaty, Anna Kiseleva, Audrey C.A. Cleuren, Kart Tomberg, Martijn A. van der Ent, Peter Gergics, K. Sue O Shea, Thomas L. Saunders, David Ginsburg
E93 expression and links to the juvenile hormone in hemipteran mealybugs with insights on female neoteny
Isabelle M Vea, Sayumi Tanaka, Tomohiro Tsuji, Takahiro Shiotsuki, Akiya Jouraku, Chieka Minakuchi
| Morphogenesis & mechanics
Anisotropic Crb accumulation, modulated by Src42A, orients epithelial tube growth in Drosophila
Ivette Olivares-Castineira, Marta Llimargas
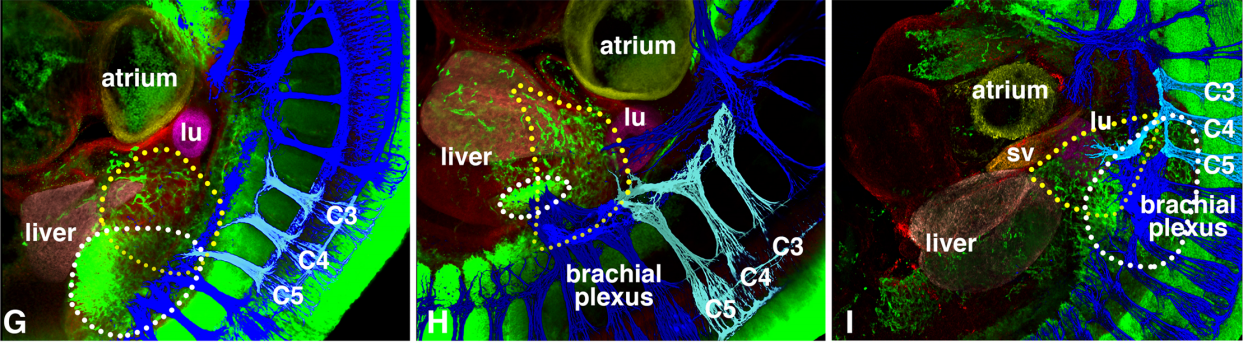
Developmental origin and morphogenesis of the diaphragm, an essential mammalian muscle
Elizabeth M Sefton, Mirialys Gallardo, Gabrielle Kardon
Spatial and temporal PCP protein dynamics coordinate cell intercalation during neural tube closure
Mitchell Butler, John B Wallingford
The nidogen-domain containing protein DEX-1 is required for epidermal remodeling in C. elegans dauers
Kristen Flatt, Caroline Beshers, Cagla Unal, Nathan Schroeder
Intermediate filaments EXC-2 and IFA-4 Maintain Luminal Structure of the Tubular Excretory Canals in Caenorhabditis elegans
HIkmat Imad Al-Hashimi, David H. Hall, Brian D Ackley, Erik A. Lundquist, Matthew Buechner
Bayesian Detection of Abnormal ADS in Mutant Caenorhabditis elegans Embryos
Wei Liang, Yuxiao Yang, Yusi Fang, Zhongying Zhao, Jie Hu
Retinal Ganglion Cell Axon Sorting at the Optic Chiasm Requires Dystroglycan
Reena Clements, Kevin M Wright
Protocadherin-mediated cell repulsion controls the central topography and efferent projections of the abducens nucleus
Kazuhide Asakawa, Koichi Kawakami
4D imaging reveals stage dependent random and directed cell motion during somite morphogenesis
James McColl, Gi Fay Mok, Anna H Lippert, Alex Ponjavic, Leila Muresan, Andrea Munsterberg
Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly
Hiroshi Yamaguchi, Toshiyuki Oda, Masahide Kikkawa, Hiroyuki Takeda
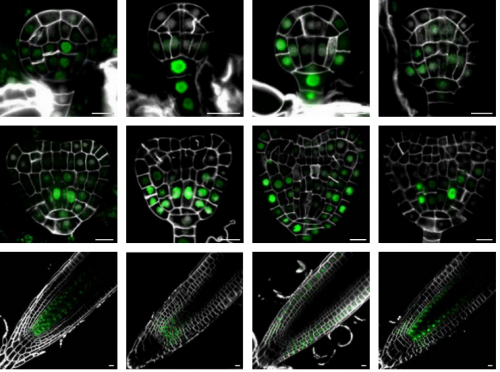
IQD proteins integrate auxin and calcium signaling to regulate microtubule dynamics during Arabidopsis development
Jos Wendrich, Bao-Jun Yang, Pieter Mijnhout, Hong-Wei Xue, Bert De Rybel, Dolf Weijers
OsIQD14 regulates rice grain shape through modulating the microtubule cytoskeleton
Bao-Jun Yang, Jos R. Wendrich, Bert De Rybel, Dolf Weijers, Hong-Wei Xue
A novel cell segmentation method for developing embryos using machine learning
Rikifumi Ota, Takahiro Ide, Tatsuo Michiue
| Genes & genomes
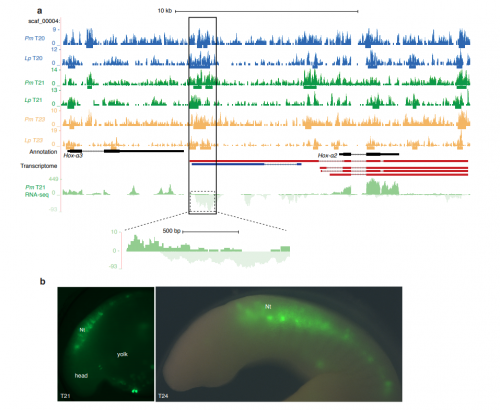
A genome-wide assessment of the ancestral neural crest gene regulatory network
Dorit Hockman, Vanessa Chong-Morrison, Daria Gavriouchkina, Stephen Green, Chris Amemiya, Jeramiah J Smith, Marianne Bronner, Tatjana Sauka-Spengler
A post-transcriptional regulatory code for mRNA stability during the zebrafish maternal-to-zygotic transition
Charles E Vejnar, Mario Abdel Messih, Carter M Takacs, Valeria Yartseva, Panos Oikonomou, Romain Christiano, Marlon Stoeckius, Stephanie Lau, Miler T Lee, Jean-Denis Beaudoin, Hiba Darwich-Codore, Tobias C Walther, Saeed Tavazoie, Daniel Cifuentes, Antonio J Giraldez
mRNA structure dynamics identifies the embryonic RNA regulome
Jean-Denis Beaudoin, Eva Maria Novoa, Charles E Vejnar, Valeria Yartseva, Carter Takacs, Manolis Kellis, Antonio J Giraldez
Dosage sensitivity of X-linked genes in human embryonic single cells
Xiaoshu Chen, Jian-Rong Yang
Two Active X-chromosomes Modulate the Growth, Pluripotency Exit and DNA Methylation Landscape of Mouse Naive Pluripotent Stem Cells through Different Pathways
Juan Song, Adrian Janiszewski, Natalie De Geest, Lotte Vanheer, Irene Talon, Taeho Oh, Vincent Pasque
A molecular network for functional versatility of HECATE transcription factors
Christophe Gaillochet, Suraj Jamge, Froukje van der Wal, Gerco Angenent, Richard Immink, Jan U. Lohmann
P-bodies and the miRNA pathway regulate translational repression of bicoid mRNA during Drosophila melanogaster oogenesis
John M. McLaughlin, Daniel FQ Smith, Irina E. Catrina, Diana P. Bratu
Bicoid gradient formation mechanism and dynamics revealed by protein lifetime analysis
Lucía Durrieu, Daniel Kirrmaier, Tatjana Schneidt, Ilia Kats, Sarada Raghavan, Michael Knop, Timothy E Saunders, Lars Hufnagel
Novel approach to quantitative spatial gene expression uncovers genetic stochasticity in the developing Drosophila eye
Sammi Ali, Sarah Signor, Konstantin Kozlov, Sergey Nuzhdin
Embryonic gene transcription in the spiny mouse (Acomys cahirinus): an investigation into the embryonic genome activation.
Jared Mamrot, David K Gardner, Peter Temple-Smith, Hayley Dickinson
Genetically regulated human NODAL splice variants are differentially post-transcriptionally processed and functionally distinct
Scott D Findlay, Olena Bilyk, Kiefer Lypka, Andrew J Waskiewicz, Lynne-Marie Postovit
A critical role for miR-142 in alveolar epithelial lineage formation
Amit Shrestha, Carraro Gianni, Nicolas Nottet, Ana Ivonne Vasquez-Armendariz, Susanne Herold, Julio Cordero, Indra Bahadur Singh, Jochen Wilhelm, Guillermo Barreto, Cho-Ming Chao, Elie El Agha, Bernard Mari, Jin-San Zhang, Saverio Bellusci
Small non-coding RNA expression in developing mouse nephron progenitor cells
Yu Leng Phua, Andrew Clugston, Kevin Hong Chen, Dennis Kostka, Jacqueline Ho
Temporal Control of Transcription by Zelda in living Drosophila embryos
Jeremy Dufourt, Antonio Trullo, Jennifer Hunter, Carola Fernandez, Jorge Lazaro, Matthieu Dejean, Lucas Morales, Katharine N Schulz, Melissa M Harrison, Ovidiu Radulescu, Cyril Favard, Mounia Lagha
MicroRNA-202 (miR-202) controls female fecundity by regulating medaka oogenesis
Stephanie Gay, Jerome Bugeon, Amine Bouchareb, Laure Henry, Jerome Montfort, Aurelie Le Cam, Julien Bobe, Violette Thermes
Lost in translation: egg transcriptome reveals molecular signature to predict developmental success and novel maternal-effect genes
Caroline T Cheung, Thaovi Nguyen, Aurelie Le Cam, Amelie Patinote, Laurent Journot, Christelle Reynes, Julien Bobe
Transcriptome Landscape of Human Oocytes and Granulosa Cells Throughout Folliculogenesis
Yaoyao Zhang, Zhiqiang Yan, Qingyuan Qing, Vicki Nisenblat, Yang Yu, Tianren Wang, Cuiling Lu, Ming Yang, Shuo Yang, Ying Yao, Xiaohui Zhu, Xi Xia, Yujiao Dang, Yixin Ren, Peng Yuan, Rong Li, Ping Liu, Hongyan Guo, Jinsong Han, Haojie He, Yu Wu, Meng Li, Kun Zhang, Yiting Wang, Jie Qiao, Jie Yan, Liying Yan
Numerous recursive sites contribute to accuracy of splicing of long introns in flies
Athma A Pai, Joseph Paggi, Karen Adelman, Christopher B Burge
Ldb1 mediates trans enhancement in mammals
Kevin Monahan, Adan Horta, Alice Mumbay-Wafula, Liqi Li, Yangu Zhao, Paul Love, Stavros Lomvardas
Down-regulated Long Noncoding RNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in preeclampsia
Lizhou Sun, Yetao Xu, Dan Wu, Jie Liu, Zhonghua Ma, Bingqing Hui, Jing Wang, Yanzi Chen, Sailan Wang, Yifan Lian
Chromatin accessibility dynamics across C. elegans development and ageing
Jurgen Janes, Yan Dong, Michael Schoof, Jacques Serizay, Alex Appert, Chiara Cerrato, Carson Woodbury, Ron Chen, Carolina Gemma, Ni Huang, Djem Kissiov, Przemyslaw Stempor, Annette Steward, Eva Zeiser, Sascha Sauer, Julie Ahringer
The tumor suppressor BRCA1/BARD1 complex localizes to the synaptonemal complex and regulates recombination under meiotic dysfunction in Caenorhabditis elegans
Qianyan Li, Takamune T Saito, Alison J Deshong, Marina Martinez Garcia, Saravanapriah Nadarajan, Katherine S Lawrence, Paula M Checchi, Monica P Colaiacovo, JoAnne Engebrecht
The BRCA1-BARD1 complex associates with the synaptonemal complex and pro-crossover factors and influences RAD-51 dynamics during Caenorhabditis elegans meiosis
Eva Janisiw, Maria Rosaria Dello Stritto, Verena Jantsch, Nicola Silva
Previously characterized Caenorhabditis elegans RNA interference-resistant mutants retain substantial silencing capacity
Jennifer L Watts, Jason S Watts, James Dalelio, Quentin Guenther
Synaptonemal complex architecture facilitates the chromosome-specific regulation of recombination in Drosophila
Cori Cahoon, G. Matthew Heenan, Zulin Yu, Jay R Unruh, Sean McKinney, R. Scott Hawley
Evolutionary analysis implicates RNA polymerase II pausing and chromatin structure in nematode piRNA biogenesis
Toni Beltran, Consuelo Barruso, Timothy Birkle, Lewis Stevens, Hillel T Schwartz, Paul Sternberg, Helene Fradin, Kristin Gunsalus, Fabio Piano, Enrique Martinez-Perez, Mark Blaxter, Peter Sarkies
The conserved phosphatase GSP-2/PP1 promotes germline immortality via small RNA-mediated genome silencing during meiosis
Katherine K Billmyre, Anna-lisa Doebley, Bree Heestand, Tony Belicard, Aya Sato-Carlton, Stephane Flibotte, Matt Simon, Megan Gnazzo, Ahna Skop, Donald Moerman, Peter Mark Carlton, Peter Sarkies, Shawn Ahmed
Transgenerational sterility of small RNA genome silencing mutants in response to germ granule dysfunction
Katherine Kretovich Billmyre, Bree Heestand, Maya Spichal, Shawn Ahmed
Epigenetic inheritance of telomere length in wild birds
Christina Bauch, Jelle J. Boonekamp, Peter Korsten, Ellis Mulder, Simon Verhulst
QTL mapping of natural variation reveals that the developmental regulator bruno reduces tolerance to P-element transposition in the Drosophila female germline
Erin Kelleher, Jaweria Jaweria, Uchechukwu Akoma, Lily Ortega, Wenpei Tang
The lysine demethylase dKDM2 is non-essential for viability, but regulates circadian rhythms in Drosophila
Yani Zheng, Yongbo Xue, Xingjie Ren, Xiao-Jun Xie, Mengmeng Liu, Yu Jia, Xiao Li, Ye Niu, Jian-Quan Ni, Yong Zhang, Jun-Yuan Ji
Nonparametric Bayesian inference of transcriptional branching and recombination identifies regulators of early human germ cell development
Christopher Andrew Penfold, Anastasiya Sybirna, John Reid, Yun Huang, Lorenz Wernisch, Zoubin Ghahramani, Murray Grant, M. Azim Surani
| Stem cells, regeneration & disease modelling
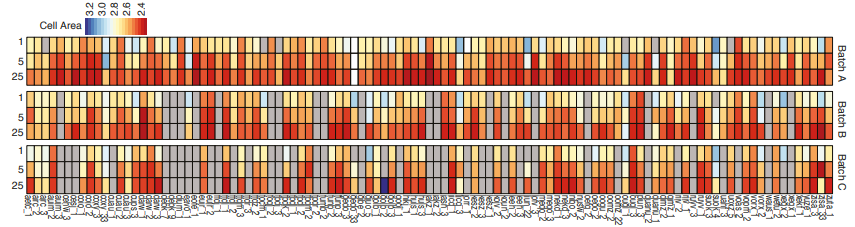
Identifying the genetic basis of variation in cell behaviour in human iPS cell lines from healthy donors
Alessandra Vigilante, Anna Laddach, Nathalie Moens, Ruta Meleckyte, Andreas Leha, Arsham Ghahramani, Oliver J. Culley, Annie Kathuria, Chloe Hurling, Alice Vickers, Mukul Tewary, Peter Zandstra, HipSci Consortium, Richard Durbin, Franca Fraternali, Oliver Stegle, Ewan Birney, Nicholas M Luscombe, Davide Danovi, Fiona M Watt
Profiling of pluripotency factors in individual stem cells and early embryos
Sarah J Hainer, Ana Boskovic, Oliver J Rando, Thomas G Fazzio
Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity
Nelly Olova, Daniel J Simpson, Riccardo Marioni, Tamir Chandra
Distinct and stage-specific contributions of TET1 and TET2 to stepwise cytosine oxidation in the transition from naive to primed pluripotency
Christopher B Mulholland, Franziska R Traube, Edris Parsa, Eva-Maria Eckl, Maximillian Schoenung, Miha Modic, Michael D Bartoschek, Paul Stolz, Joel Ryan, Thomas Carell, Heinrich Leonhardt, Sebastian Bultmann
Transcriptional repression by FACT is linked to regulation of chromatin accessibility at the promoter of ES cells
Peter Tessarz, Constantine Mylonas
Acidic microenvironment shaped by lactate accumulation promotes pluripotency through multiple mechanisms
Wen-Ting Guo, Shao-Hua Wang, Xiao-Shan Zhang, Ming Shi, Jing Hao, Xi-Wen Wang, Kai-Li Gu, Fei-Fei Duan, Ying Yan, Xi Yang, Chao Zhang, Le-Qi Liao, Yangming Wang
Chromosome compartmentalization replacement during stem cell differentiation
Yuri Eidelman, Svetlana Slanina, Valentina Pyatenko, Sergey Andreev
Coupling initial hydrogel microenvironments with mesenchymal stem cell remodeling to define a dynamic hematopoietic stem cell niche
Aidan E Gilchrist, Sunho Lee, Yuhang Hu, Brendan Harley
Longitudinal single cell fate in hematopoiesis in vivo using cellular barcoding and DiSNE movie visualization
Jerry Gao, Dawn Lin, Edmund Crampin, Shalin Naik
Urothelial organoids originate from Cd49f-High stem cells and display Notch-dependent differentiation capacity
Francisco X Real, Catarina P Santos, Eleonora Lapi, Laura Alvaro-Espinosa, Asuncion Fernandez-Barral, Antonio Barbachano, Diego Megias, Alberto Munoz
Onset of differentiation is posttranscriptionally controlled in adult neural stem cells
Avni Baser, Yonglong Dang, Maxim Skabkin, Guelce Guelcueler Balta, Georgios Kalamakis, Susanne Kleber, Manuel Goepferich, Roman Schefzik, Alejandro Santos Lopez, Enric Llorens Bobadilla, Carsten Schultz, Bernd Fischer, Ana Martin-Villalba
Single cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors
Nicole AJ Krentz, Michelle Lee, Eric E Xu, Shugo Sasaki, Francis C Lynn
A Comparative Assessment of Human and Chimpanzee iPSC-derived Cardiomyocytes with Primary Heart Tissues
Bryan J Pavlovic, Lauren E Blake, Julien Roux, Claudia Chavarria, Yoav Gilad
Temporal epigenomic profiling identifies AHR and GLIS1 as super-enhancer controlled regulators of mesenchymal multipotency
Deborah Gerard, Florian Schmidt, Aurelien Ginolhac, Martine Schmitz, Rashi Halder, Peter Ebert, Marcel H. Schulz, Thomas Sauter, Lasse Sinkkonen
Correlated random walks of human embryonic stem cell in-vitro
L E Wadkin, S Orozco-Fuentes, I Neganova, G Swan, A Laude, M Lako, A Shukurov, N G Parker
An Epiblast Stem Cell derived multipotent progenitor population for axial extension
Shlomit Edri, Penelope Hayward, Peter Baillie-Johnson, Benjamin Steventon, Alfonso Martinez Arias
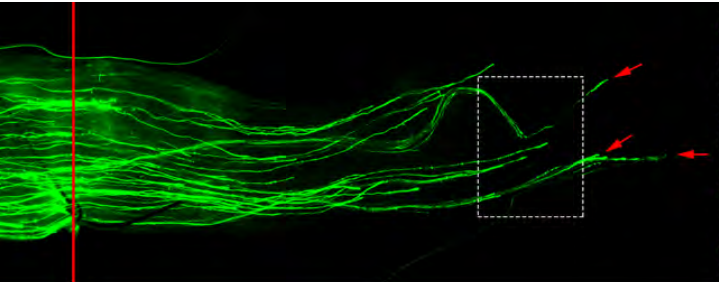
Novel role of Lin28 signaling in regulation of mammalian PNS and CNS axon regeneration
Xue-Wei Wang, Chang-Mei Liu, Philip A Hall, Jing-Jing Jiang, Christopher D Katchis, Sehwa Kang, Bryan C Dong, Shuxin Li, Feng-Quan Zhou
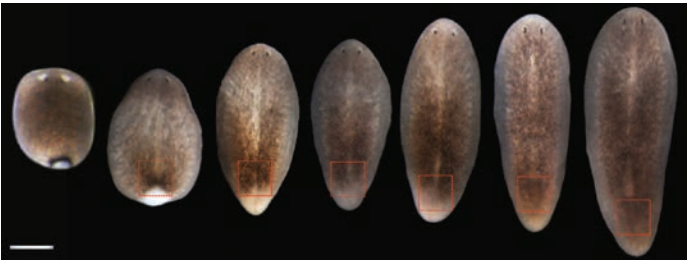
Positional information specifies the site of organ regeneration and not tissue maintenance in planarians
Eric M. Hill, Christian P Petersen
GABA promotes survival and axonal regeneration in identifiable descending neurons after spinal cord injury in larval lampreys
Daniel Romaus-Sanjurjo, Rocio Ledo-Garcia, Blanca Fernandez-Lopez, Kendra Hanslik, Jennifer R Morgan, Anton Barreiro-Iglesias, Maria Celina Rodicio
Experimentally Induced Metamorphosis in Axolotl (Ambystoma mexicanum) Under Constant Diet Restructures Microbiota Accompanied by Reduced Limb Regenerative Capacity
Turan Demircan, Guvanch Ovezmyradov, Berna YILDIRIM, Iknur Keskin, Ayse Elif ilhan, Ece Cana Fescioglu, Gurkan Ozturk, Suleyman Yildirim
Capn5 expression in the healthy and regenerating zebrafish retina
Cagney E. Coomer, Ann C Morris
A 3D model of human skeletal muscle innervated with stem cell-derived motor neurons enables epsilon-subunit targeted myasthenic syndrome studies
Mohsen Afshar Bakooshli, Ethan S Lippmann, Ben Mulcahy, Kayee Tung, Elena Pegoraro, Henry Ahn, Howard Ginsberg, Mei Zhen, Randolph S Ashton, Penney M Gilbert
Aneuploidy Promotes Intestinal Dysplasia in Drosophila
Luis P Resende, Augusta Monteiro, Rita Bras, Tatiana Lopes, Claudio E Sunkel
All-optical electrophysiology for high-throughput functional characterization of human iPSC-derived motor neuron model of ALS
Evangelos Kiskinis, Joel M Kralj, Peng Zou, Eli N Weinstein, Hongkang Zhang, Konstantinos Tsioras, Ole Wiskow, J. Alberto Ortega, Kevin Eggan, Adam E Cohen
Transcriptional and electrophysiological aberrations in an induced pluripotent stem cell-derived model of spinocerebellar ataxia type 7
Lauren M Watson, Danielle C Smith, Joseph V Raimondo, Richard J Burman, Robea Ballo, Janine Scholefield, Lyn Tyers, Sally A Cowley, Matthew JA Wood, Susan H Kidson, Leslie Jacquie Greenberg
Evo-devo & evo
Adipose fin development and its relation to the evolutionary origins of median fins
Thomas Stewart, Robert Ho, Melina A Hale
Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution
Sebastian Kittelmann, Alexandra D. Buffry, Franziska A. Franke, Isabel Almudi, Marianne Yoth, Gonzalo Sabaris, Juan Pablo Couso, Maria D. S. Nunes, Nicolas Frankel, Jose Luis Gomez-Skarmeta, Jose Pueyo-Marques, Saad Arif, Alistair P. McGregor
The X chromosome of the German cockroach, Blattella germanica, is homologous to a fly X chromosome despite 400 million years divergence
Richard P Meisel, Judith R Wexler
The Hedgehog signaling domain was acquired from a prokaryote.
Henk Roelink
Evolution of the D. melanogaster chromatin landscape and its associated proteins
Elise Parey, Anton Crombach
Thirty clues to the exceptional diversification of flowering plants
Susana Magallon, Luna Sanchez-Reyes, Sandra Gomez Acevedo
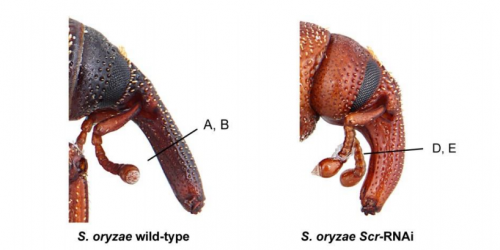
Developmental genetics in a complex adaptive structure, the weevil rostrum
Steven Davis
Germ layer specific regulation of cell polarity and adhesion gives insight into the evolution of mesoderm.
Miguel Salinas-Saavedra, Amber Q. Rock, Mark Q. Martindale
Placozoa and Cnidaria are sister taxa
Christopher E. Laumer, Harald Gruber-Vodicka, Michael G. Hadfield, Vicki B. Pearse, Ana Riesgo, John C. Marioni, Gonzalo Giribet
Functional Characterization of Enhancer Evolution in the Primate Lineage
Jason Chesler Klein, Aidan Keith, Vikram Agarwal, Timothy Durham, Jay Shendure
Tempo, mode, and fitness effects of mutation in Caenorhabditis elegans over 400 generations of minimal selection
Ayush Shekhar Saxena, Matthew P Salomon, Chikako Matsuba, Shu-Dan Yeh, Charles F Baer
Transposable elements and gene expression during the evolution of amniotes
Lu Zeng, Stephen Pederson, Dan Kortschak, David Adelson
Diverse RNA interference strategies in early-branching metazoans
Andrew D Calcino, Selene L Fernandez-Valverde, Ryan J Taft, Bernard M Degnan
What is an archaeon and are the Archaea really unique?
Ajith Harish
CHARACTERIZATION OF THE bHLH FAMILY OF TRANSCRIPTIONAL REGULATORS IN THE ACOEL S. roscoffensis AND THEIR PUTATIVE ROLE IN NEUROGENESIS
Elena Perea-Atienza, Simon G. Sprecher, Pedro Martinez
Multiple large-scale gene and genome duplications during the evolution of hexapods
Zheng Li, George Tiley, Sally Galuska, Chris Reardon, Thomas Kidder, Rebecca Rundell, Michael S. Barker
Finding Nemo’s Genes: A chromosome-scale reference assembly of the genome of the orange clownfish Amphiprion percula
Robert Lehmann, Damien J Lightfoot, Celia Schunter, Craig T Michell, Hajime Ohyanagi, Katsuhiko Mineta, Sylvain Foret, Michael Berumen, David J Miller, Manuel Aranda, Takashi Gojobori, Philip L Munday, Timothy Ravasi
Improved genome assembly and annotation for the rock pigeon (Columba livia)
Carson Holt, Michael Campbell, David Keays, Nathaniel Edelman, Aurelie Kapusta, Emily Maclary, Eric Domyan, Alexander Suh, Wesley Warren, Mark Yandell, Thomas Gilbert, Michael D. Shapiro
A phylogenomic framework and timescale for comparative studies of tunicates
Frederic Delsuc, Herve Philippe, Georgia Tsagkogeorga, Paul Simion, Marie-Ka Tilak, Xavier Turon, Susanna Lopez-Legentil, Jacques Piette, Patrick Lemaire, Emmanuel J. P. Douzery
Natural selection on gene-specific codon usage bias is common across eukaryotes
Zhen Peng, Hani Zaher, Yehuda Ben-Shahar
Cell biology
Spatial Control of Draper Receptor Signaling Initiates Apoptotic Cell Engulfment
Adam P. Williamson, Ronald D. Vale
Assembly of a Parts List of the Human Mitotic Cell Cycle Machinery
Bruno Giotti, Sz-Hau Chen, Mark W Barnett, Tim Regan, Tony Ly, Stefan Wiemann, David A Hume, Tom C Freeman
CDC20B is required for deuterosome-mediated centriole production in multiciliated cells
Diego R. Revinski, Laure-Emmanuelle Zaragosi, Camille Boutin, Sandra Ruiz-Garcia, Marie Deprez, Virginie Thome, Olivier Rosnet, Anne-Sophie Gay, Olivier Mercey, Agnes Paquet, Nicolas Pons, Gilles Ponzio, Brice Marcet, Laurent Kodjabachian, Pascal Barbry
Scaffolding of RhoA contractile signaling by anillin: a regulatory analogue of kinetic proofreading
Srikanth Budnar, Kabir B. Husain, Guillermo A. Gomez, Maedeh Naghibosidat, Suzie Verma, Nicholas A. Hamilton, Richard G. Morris, Alpha S. Yap
A mechanosensitive RhoA pathway that protects epithelia against acute tensile stress.
Bipul R. Acharya, Alexander Nestor-Bergmann, Xuan Liang, Srikanth Budnar, Oliver E. Jensen, Zev Bryant, Alpha S. Yap
Dynamics of sister chromatid resolution during cell cycle progression
Rugile Stanyte, Johannes Nuebler, Claudia Blaukopf, Rudolf Hoefler, Roman Stocsits, Jan-Michael Peters, Daniel W Gerlich
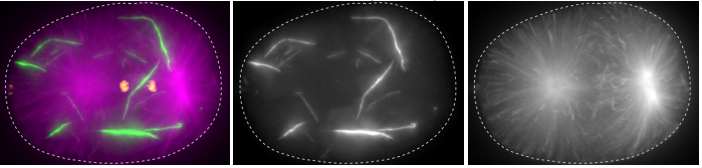
Self-assembly of the RZZ complex into filaments drives kinetochore expansion in the absence of microtubule attachment
Reto Gassmann, Cláudia Pereira, Rita M Reis, José B Gama, Dhanya K Cheerambathur, Ana X Carvalho
Dynamic Kinetochore Size Regulation Promotes Microtubule Capture And Chromosome Biorientation In Mitosis
Carlos Sacristan, Misbha Ahmad, Jenny Keller, Job Fermie, Vincent Groenewold, Eelco Tromer, Alexander Fish, Roberto Melero, Jose Maria Carazo, Judith Klumperman, Andrea Musacchio, Anastassis Perrakis, Geert JPL Kops
Optogenetic reconstitution reveals that Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble
Masako Okumura, Toyoaki Natsume, Masato T Kanemaki, Tomomi Kiyomitsu
A novel microtubule nucleation pathway for meiotic spindle assembly in oocytes
Pierre ROME, Hiroyuki OHKURA
A compartmentalized signaling network mediates crossover control in meiosis
Liangyu Zhang, Simone Koehler, Regina Rillo-Bohn, Abby F. Dernburg
Microtubule end tethering of a processive Kinesin-8 motor Kif18b is required for spindle positioning
Toni McHugh, Agata Gluszek-Kustusz, Julie Welburn
Mitotic progression, arrest, exit or death is determined by centromere integrity and independent of de novo transcription
Marco Cruz, Maria Alba Fernandez, Wilfred van Ijcken, Niels Galjart, Arockia Jeyaprakash Arulanandam, Helder Maiato, Cristina Ferras
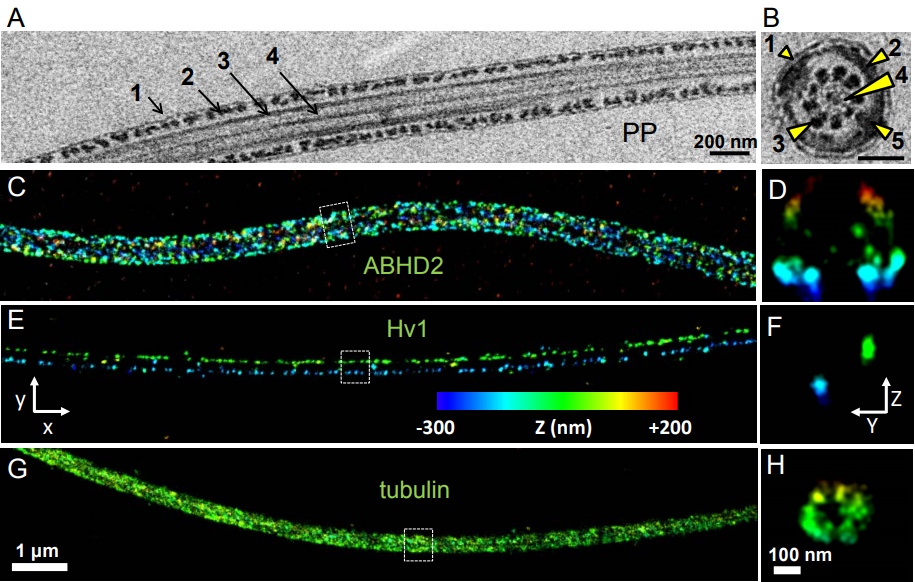
Asymmetrically Positioned Flagellar Control Units Regulate Human Sperm Rotation
Melissa Miller, Samuel Kenny, Nadja Mannowetz, Steven Mansell, Michal Wojcik, Sarah Mendoza, Robert Zucker, Ke Xu, Polina V Lishko
Regulation of DVL2 ubiquitylation by USP9X biases participation in canonical or non-canonical WNT signalling
Casey P Nielsen, Kristin K Jernigan, Jason A. MacGurn
Spatiotemporal manipulation of ciliary glutamylation reveals its roles in intraciliary trafficking and Hedgehog signaling
Shi-Rong Hong, Cuei-Ling Wang, Yao-Shen Huang, Yu-Chen Chang, Ya-Chu Chang, Ganesh V. Pusapati, Chun-Yu Lin, Ning Hsu, Hsiao-Chi Cheng, Yueh-Chen Chiang, Wei-En Huang, Nathan C. Shaner, Rajat Rohatgi, Takanari Inoue, Yu-Chun Lin
Cytoplasmic sequestration of the RhoA effector mDiaphanous1 by Prohibitin2 promotes muscle differentiation
Jyotsna Dhawan, Amena Saleh, Gunasekaran Subramaniam, Swasti Raychaudhuri
Polyamines support myogenesis by facilitating myoblast migration
Shirley Brenner, Yulia Feiler, Chaim Kahana
Stabilising and destabilising kinesin complexes queue at plus tips to ensure microtubule catastrophe at cell ends
John C Meadows, Liam J Messin, Anton Kamnev, Theresa C Lancaster, Mohan K Balasubramanian, Robert A Cross, Jonathan B.A. Millar
CLASP promotes microtubule array reorientation by acting as a specific severing rescue factor
Jelmer J Lindeboom, Masayoshi Nakamura, Marco Saltini, Anneke Hibbel, Ankit Walia, Tijs Keetlaar, Anne Mie C Emons, John C Sedbrook, Viktor Kirik, Bela M Mulder, David W Ehrhardt
Three-dimensional cell geometry controls excitable membrane signaling in Dictyotelium cells
Marcel Hörning, Tatsuo Shibata
An experimental and computational framework to build a dynamic protein atlas of human cell division
Yin Cai, M. Julius Hossain, Jean-Karim Heriche, Antonio Z. Politi, Nike Walther, Birgit Koch, Malte Wachsmuth, Bianca Nijmeijer, Moritz Kueblbeck, Marina Martinic Kavur, Rene Ladurner, Jan-Michael Peters, Jan Ellenberg
Modelling
Bradly J Alicea, Richard Gordon
DevoWorm: data-theoretical synthesis of C. elegans development.
Bradly John Alicea, Richard Gordon, Thomas E. Portegys

Beyond activator-inhibitor networks: the generalised Turing mechanism
Stephen Smith, Neil Dalchau
Selection for synchronized cell division in simple multicellular organisms
Jason Olejarz, Kamran Kaveh, Carl Veller, Martin A. Nowak
Branch-recombinant Gaussian processes for analysis of perturbations in biological time series
Christopher A Penfold, Anastasiya Sybirna, John Reid, Yun Huang, Lorenz Wernisch, Zoubin Ghahramani, Murray Grant, M Azim Surani
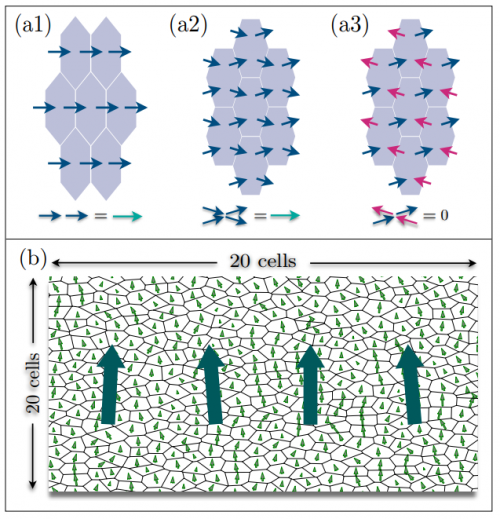
The Role of Cytoplasmic Interactions in the Collective Polarization of Tissues and its Interplay with Cellular Geometry
Shahriar Shadkhoo, Madhav Mani
Interplay between epidermal stem cell dynamics and dermal deformations
Yasuaki Kobayashi, Yusuke Yasugahira, Hiroyuki Kitahata, Mika Watanabe, Ken Natsuga, Masaharu Nagayama
Confluent and non-confluent phases in a model of cell tissue
Eial Teomy, David A. Kessler, Herbert Levine
The Role of Cytoplasmic Interactions in the Collective Polarization of Tissues and its Interplay with Cellular Geometry
Shahriar Shadkhoo, Madhav Mani
Optimal feedback mechanisms for regulating cell numbers
Saurabh Modi, Abhyudai Singh
Transition from actin-driven to water-driven cell migration depends on external hydraulic resistance
Yizeng Li, Sean X. Sun
Tools & resources
| Imaging etc.
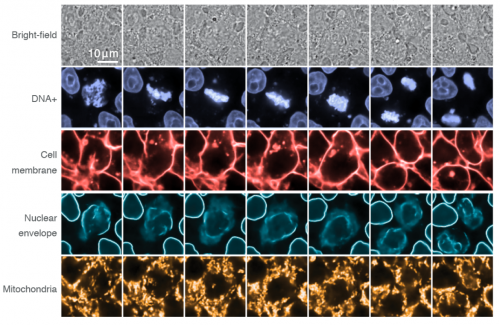
Label-free prediction of three-dimensional fluorescence images from transmitted light microscopy
Chawin Ounkomol, Sharmishtaa Seshamani, Mary M Maleckar, Forrest Collman, Gregory Johnson
Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust
Harry M.T. Choi, Maayan Schwarzkopf, Mark E. Fornace, Aneesh Acharya, Georgios Artavanis, Johannes Stegmaier, Alexandre Cunha, Niles A. Pierce
Automated sequential chromogenic IHC double staining with two HRP substrates.
Kenneth H Petersen, Jesper Lohse, Lasse Ramsgaard
Forget Pixels: Adaptive Particle Representation of Fluorescence Microscopy Images
Bevan L. Cheeseman, Ulrik Günther, Mateusz Susik, Krzysztof Gonciarz, Ivo F. Sbalzarini
Label-free assessment of pre-implantation embryo quality by the Fluorescence Lifetime Imaging Microscopy (FLIM)-phasor approach
Ning Ma, Nabora Reyes de Mochel, Paula Pham, Tae Yoo, Ken Cho, Michelle Digman
Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system
Yuriko Kakimoto, Shinya Tashiro, Rieko Kojima, Yuuki Morozumi, Toshiya Endo, Yasushi Tamura
Signal-to-Noise ratio made easy: A tool to assess your confocal performance
Alexia Ferrand, Kai D Schleicher, Nikolaus Ehrenfeuchter, Wolf Heusermann, Oliver Biehlmaier
Smart Region-Growing: a novel algorithm for the segmentation of 3D clarified confocal image stacks
Alejandro Callara, Chiara Magliaro, Arti Ahluwalia, Nicola Vanello
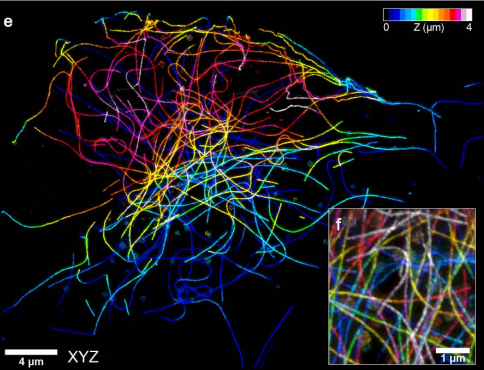
Expansion stimulated emission depletion microscopy (ExSTED)
Mengfei Gao, Riccardo Maraspini, Oliver Beutel, Amin Zehtabian, Britta Eickholt, Alf Honigmann, Helge Ewers
Complementary studies of lipid membrane dynamics using iSCAT and STED Fluorescence Correlation Spectroscopy
Francesco Reina, Silvia Galiani, Dilip Shrestha, Erdinc Sezgin, Gabrielle de Wit, Daniel Cole, B. Christoffer Lagerholm, Philipp Kukura, Christian Eggeling
Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy
Mohammed Mahamdeh, Steve Simmert, Anna Luchniak, Erik Schaeffer, Jonathon Howard
A new approach to design artificial 3D micro-niches with combined chemical, topographical and rheological cues
Celine Stoecklin, Zhang Yue, Wilhelm W. Chen, Richard de Mets, Eileen Fong, Vincent Studer, Virgile Viasnoff
| Genome tools
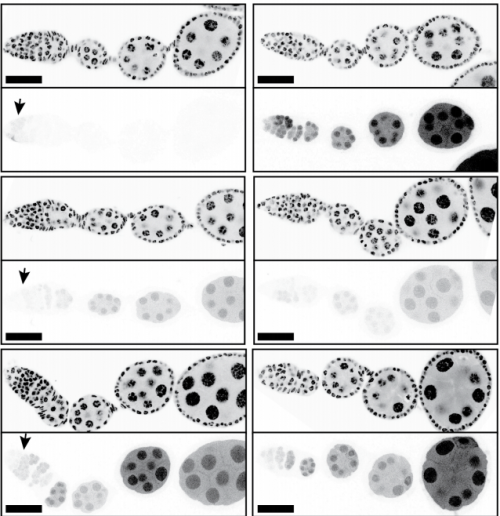
Efficient expression of genes in the Drosophila germline using a UAS-promoter free of interference by Hsp70 piRNAs
Steven Z DeLuca, Allan C Spradling
Germline silencing of UASt depends on the piRNA pathway
Yi-Chun Huang, Henry Moreno, Sarayu Row, Dongyu Jia, Wu-Min Deng
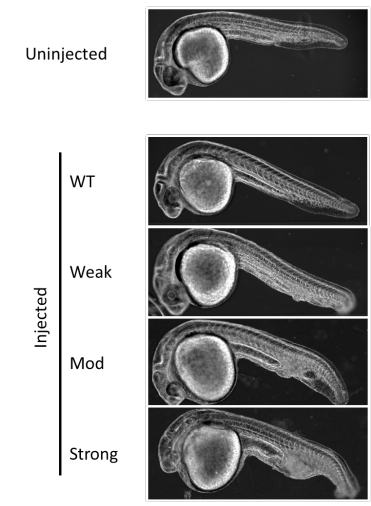
Toward Precision Molecular Surgery: Robust, Selective Induction of Microhomology-mediated End Joining in vivo
Hirotaka Ata, Thomas L Ekstrom, Gabriel Martinez-Galvez, Carla M Mann, Alexey V Dvornikov, Kyle J Schaefbauer, Alvin C Ma, Drena Dobbs, Karl J Clark, Stephen C Ekker
Corrigendum and follow-up: Whole genome sequencing of multiple CRISPR-edited mouse lines suggests no excess mutations.
Kellie A. Schaefer, Benjamin W. Darbro, Diana F. Colgan, Stephen H. Tsang, Alexander G. Bassuk, Vinit B. Mahajan
A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 nucleases in rice
Xu Tang, Guanqing Liu, Jianping Zhou, Qiurong Ren, Qi You, Li Tian, Xuhui Xin, Zhaohui Zhong, Binglin Liu, Xuelian Zheng, Dengwei Zhang, Aimee Malzahn, Zhiyun Gong, Yiping Qi, Tao Zhang, Yong Zhang
CRISPR-Tag: an Efficient DNA Tagging System in Living Cells
Baohui Chen, Wei Zou, Bo Huang
A homing CRISPR mouse resource for barcoding and lineage tracing
Reza Kalhor, Kian Kalhor, Kathleen Leeper, Amanda Graveline, Prashant Mali, George M Church
High-throughput antibody engineering in mammalian cells by CRISPR/Cas9-mediated homology-directed mutagenesis
Derek M Mason, Cedric R Weber, Cristina Parola, Simon M Meng, Victor Greiff, William J Kelton, Sai T Reddy
An optimized electroporation approach for efficient CRISPR/Cas9 genome editing in murine zygotes
Simon E. Tröder, Lena K. Ebert, Linus Butt, Sonja Assenmacher, Bernhard Schermer, Branko Zevnik
Ultra-parallel ChIP-seq by barcoding of intact nuclei
Laura Arrigoni, Hoor Al-Hasani, Fidel Ramirez, Ilaria Panzeri, Devon P Ryan, Diana Santacruz, Nadia Kress, Andrew J Pospisilik, Ulrike Boenisch, Thomas Manke
Generation and validation of homozygous fluorescent knock-in cells using genome editing
Birgit Koch, Bianca Nijmeijer, Moritz Kueblbeck, Yin Cai, Nike Walther, Jan Ellenberg
A comprehensive toolkit to enable MinION long-read sequencing in any laboratory
Miriam Schalamun, David Kainer, Eleanor Beavan, Ramawatar Nagar, David Eccles, John Rathjen, Robert Lanfear, Benjamin Schwessinger
Topographer Reveals Dynamic Mechanisms of Cell Fate Decisions from Single-Cell Transcriptomic Data
Jiajun Zhang, Tianshou Zhou, Qing Nie
A biological-computational human cell lineage discovery platform based on duplex molecular inversion probes
Liming Tao, Ofir Raz, Zipora Marx, Tamir Biezuner, Shiran Amir, Lilach Milo, Rivka Adar, Amos Onn, Noa Chapal-Ilani, Veronika Berman, Ron Levy, Barak Oron, Ehud Shapiro
Towards Improving Embryo Selection: Simultaneous Next Generation Sequencing Of DNA And RNA From A Single Trophectoderm Biopsy
Noga Fuchs Weizman, Brandon Wyse, Ran Antes, Zenon Ibarrientos, Mugundhine Sangaralingam, Gelareh Motamedi, Valeriy Kuznyetsov, Svetlana Madjunkova, Clifford L Librach
NanoMod: a computational tool to detect DNA modifications using Nanopore long-read sequencing data
Qian Liu, Daniela C. Georgieva, Dieter Egli, Kai Wang
Auxin-Mediated Sterility Induction System for Longevity and Mating Studies in Caenorhabditis elegans
Katja R Kasimatis, Megan J. Moerdyk-Schauecker, Patrick C. Phillips
DECODE-ing sparsity patterns in single-cell RNA-seq
Shahin Mohammadi, Jose Davila-Velderrain, Manolis Kellis, Ananth Grama
Take ACTION to characterize the functional identity of single cells
Shahin Mohammadi, Vikram Ravindra, David Gleich, Ananth Grama
Pisces: An Accurate and Versatile Variant Caller for Somatic and Germline Next-Generation Sequencing Data
Tamsen Dunn, Gwenn Berry, Dorothea Emig-Agius, Yu Jiang, Serena Lei, Anita Iyer, Nitin Udar, Han-Yu Chuang, Jeff Hegarty, Michael Dickover, Brandy Klotzle, Justin Robbins, Marina Bibikova, Marc Peeters, Michael Stromberg
Research practice & education
Twenty steps towards an adequate inferential interpretation of p-values
Norbert Hirschauer, Sven Grüner, Oliver Mußhoff, Claudia Becker
Adjutant: an R-based tool to support topic discovery for systematic and literature reviews
Anamaria Crisan, Tamara Munzner, Jennifer L. Gardy
Quantifying the impact of public omics data
Yasset Perez-Riverol, Andrey Zorin, Gaurhari Dass, Mihai Glonț, Juan Antonio Vizcaino, Andrew Jarnuczak, Robert Petryszak, Peipei Ping, Henning Hermjakob
Changes in Acceptance of Evolution and Associated Factors during a Year of Introductory Biology: The Shifting Impacts of Biology Knowledge, Politics, Religion, Demographics, and Understandings of the Nature of Science
Ryan D.P. Dunk, Jason R. Wiles
Persistent Underrepresentation of Women’s Science in High Profile Journals
Yiqin Alicia Shen, Jason M Webster, Yuichi Shoda, Ione Fine
Female grant applicants are equally successful when peer reviewers assess the science, but not when they assess the scientist
Holly O Witteman, Michael Hendricks, Sharon Straus, Cara Tannenbaum
A Data Citation Roadmap for Scientific Publishers
Helena Cousijn, Amye Kenall, Emma Ganley, Melissa Harrison, David Kernohan, Thomas Lemberger, Fiona Murphy, Patrick Polischuk, Simone Taylor, Maryann Martone, Timothy Clark
We need to talk about reliability: Making better use of test retest studies for study design and interpretation
Granville James Matheson
Dhanush Kumar, Rakhi Radhamani, Nijin Nizar, Krishnashree Achuthan, Bipin Nair, Shyam Diwakar
Researcher engagement in policy deemed societally beneficial yet unrewarded
Gerald G Singh, Vinicius Farjalla, Bing Chen, Andrew Pelling, Elvan Ceyhan, Martin Dominik, Eva Alisic, Jeremy Kerr, Noelle Selin, Ghada Bassioni, Elena Bennett, Andrew Kemp, Kai MA Chan
Chris HJ Hartgerink, Marino van Zelst
Why not…
An Observational Study of Ballooning in Large Spiders: Nanoscale Multi-Fibres Enable Large Spiders’ Soaring Flight
Moonsung Cho, Peter Neubauer, Christoph Fahrenson, Ingo Rechenberg
Quantifying the unquantifiable: why Hymenoptera — not Coleoptera — is the most speciose animal order
Andrew A Forbes, Robin K Bagley, Marc A Beer, Alaine C Hippee, Heather A Widmayer
Ancient DNA reveals the chronology of walrus ivory trade from Norse Greenland
Bastiaan Star, James H Barrett, Agata T Gondek, Sanne Boessenkool


 (No Ratings Yet)
(No Ratings Yet)