March in preprints
Posted by the Node, on 1 April 2019
Welcome to our monthly trawl for developmental biology (and related) preprints.
This month we found three hydra preprints, lots of developmental mechanics, a typically hearty serving of single cell transcriptomic analyses and a survey of the life of PIs.
The preprints were hosted on bioRxiv, PeerJ, and arXiv. Let us know if we missed anything, and use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice & education
Why not…
Developmental biology
| Patterning & signalling
Shh induces symmetry breaking in the presomitic mesoderm by inducing tissue shear and orientated cell rearrangements
J. Yin, T. E. Saunders
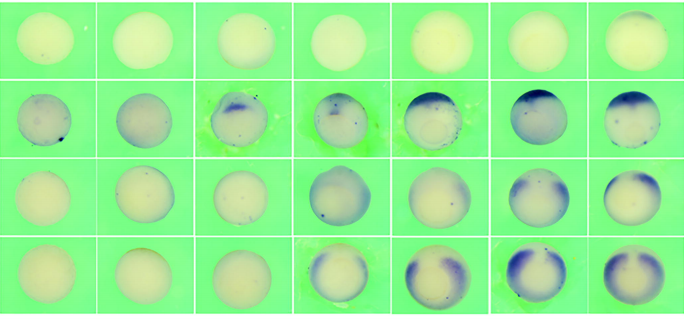
Patterning of the vertebrate head in time and space by BMP signalling
Kongju Zhu, Herman P. Spaink, Antony J. Durston
ERK1/2 signalling dynamics promote neural differentiation by regulating the polycomb repressive complex
Claudia I. Semprich, Vicki Metzis, Harshil Patel, James Briscoe, Kate G. Storey
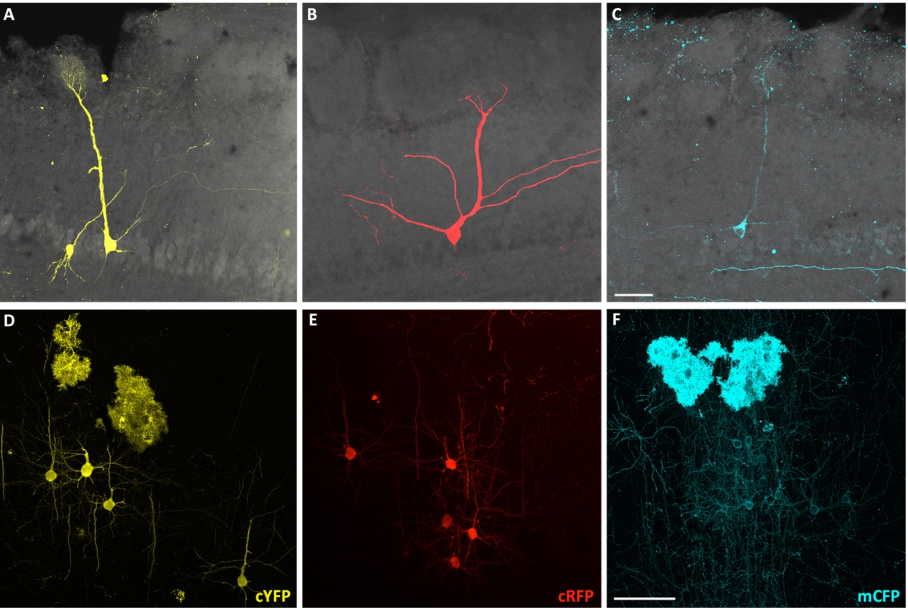
Lineage does not regulate the connectivity of projection neurons in the mouse olfactory bulb
Luis Sánchez-Guardado, Carlos Lois
Jag1 coordinates release from multipotency with cell fate choice in the developing pancreas
Philip A. Seymour, Caitlin A. Collin, Mette C. Jørgensen, Itaru Imayoshi, Kristian H. de Lichtenberg, Raphael Kopan, Ryoichiro Kageyama, Palle Serup
Pluripotent Stem Cell-derived Cerebral Organoids Reveal Human Oligodendrogenesis with Dorsal and Ventral Origins
Hyosung Kim, Ranjie Xu, Padmashri Ragunathan, Anna Dunaevsky, Ying Liu, Cheryl F. Dreyfus, Peng Jiang
FGF signalling regulates enhancer activation during ear progenitor induction
Monica Tambalo, Maryam Anwar, Mohi Ahmed, Andrea Streit
Notch signaling restricts FGF pathway activation in parapineal cells to promote their collective migration
Lu Wei, Patrick Blader, Myriam Roussigné
Pax6 and KDM5C co-occupy a subset of developmentally critical genes including Notch signaling regulators in neural progenitors
Giulia Gaudenzi, Olga Dethlefsen, Julian Walfridsson, Ola Hermanson
Microglia actively remodels adult hippocampal neurogenesis through the phagocytosis secretome
Irune Diaz-Aparicio, Iñaki Paris, Virginia Sierra-Torre, Ainhoa Plaza-Zabala, Noelia Rodríguez-Iglesias, Mar Márquez-Ropero, Sol Beccari, Oihane Abiega, Elena Alberdi, Carlos Matute, Irantzu Bernales, Angela Schulz, Lilla Otrokocsi, Beata Sperlagh, Kaisa E. Happonen, Greg Lemke, Mirjana Maletic-Savatic, Jorge Valero, Amanda Sierra
Neuronal programming by microbiota enables environmental regulation of intestinal motility
Yuuki Obata, Stefan Boeing, Álvaro Castaño, Ana Carina Bon-Frauches, Mercedes Gomez de Agüero, Werend Boesmans, Bahtiyar Yilmaz, Rita Lopes, Almaz Huseynova, Muralidhara Rao Maradana, Pieter Vanden Berghe, Andrew J. Murray, Brigitta Stockinger, Andrew J. Macpherson, Vassilis Pachnis
Timing and Duration of Gbx2 Expression Delineates Thalamocortical and Dopaminergic Medial Forebrain Bundle Circuitry
Elizabeth Normand, Catherine Browning, Mark Zervas
High Epha1 expression is a potential cell surface marker for embryonic neuro-mesodermal progenitors
Luisa de Lemos, Ana Nóvoa, Moisés Mallo
CELL SPECIFIC REACTIVATION OF EPICARDIUM AT THE ORIGIN OF FIBRO-FATTY REMODELING OF THE ATRIAL MYOCARDIUM
N Suffee-Mosbah, thomas Moore-Morris, nathalie Mougenot, gilles dilanaian, myriam berthet, bernd jagla, julie Proukhnitzky, pascal leprince, david tregouet, michel puceat, stephane hatem
SULTA4A1 modulates synaptic development and function by promoting the formation of PSD-95/NMDAR complex
Lorenza Culotta, Benedetta Terragni, Ersilia Vinci, Alessandro Sessa, Vania Broccoli, Massimo Mantegazza, Chiara Verpelli
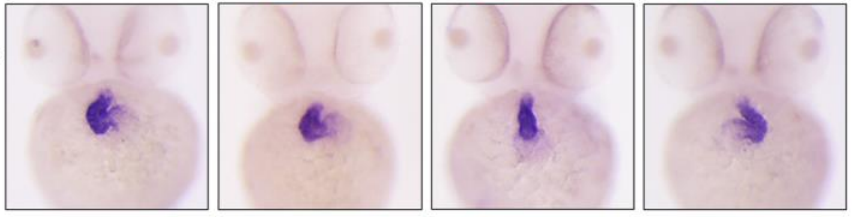
Chemokine signaling links cell cycle progression and cilia formation for left-right symmetry breaking
Jingwen Liu, Chengke Zhu, Guozhu Ning, Liping Yang, Yu Cao, Sizhou Huang, Qiang Wang
Metabolic reprogramming from glycolysis to amino acid utilization in cardiac HIF1α deficient mice
Ivan Menendez-Montes, Beatriz Escobar, Beatriz Palacios, Manuel J. Gomez, Elena Bonzon, Alessia Ferrarini, Ana Vanessa Alonso, Luis Jesus Jimenez-Borreguero, Jesus Vázquez, Silvia Martin-Puig
In the chick embryo, estrogen can induce chromosomally male ZZ left gonad epithelial cells to form an ovarian cortex which supports oogenesis
Silvana Guioli, Debiao Zhao, Sunil Nandi, Michael Clinton, Robin Lovell-Badge
CDK2 kinase activity is a regulator of male germ cell fate
Priti Singh, Ravi Patel, Nathan Palmer, Jennifer Grenier, Darius Paduch, Philipp Kaldis, Andrew Grimson, John Schimenti
Ligands, receptors and transcription factors that mediate inter-cellular and intra-cellular communication during ovarian follicle development
Beatriz Penalver Bernabe, Teresa Woodruff, Linda Broadbelt, Lonnie Shea
Sterol-O acyltransferase 1 is inhibited by gga-miR-181a-5p and gga-miR-429-3p through the TGFβ pathway in endodermal epithelial cells of Japanese quail
Han-Jen Lin, Chiao-Wei Lin, Harry J. Mersmann, Shih-Torng Ding
Refinement of the primate corticospinal pathway during prenatal development
Ana Rita Ribeiro Gomes, Etienne Olivier, Herbert P Killackey, Pascale Giroud, Michel Berland, Kenneth Knoblauch, Colette Dehay, Henry Kennedy
Preliminary evidence that maternal immune activation specifically increases diagonal domain volume in the rat brain during early postnatal development
Tobias C. Wood, Michelle E. Edye, Michael K. Harte, Joanna C. Neill, Eric P. Prinssen, Anthony C. Vernon
Human cortical organoids expose a differential function of GSK3 on direct and indirect neurogenesis
Alejandro López-Tobón, Carlo Emanuele Villa, Cristina Cheroni, Sebastiano Trattaro, Nicolò Caporale, Paola Conforti, Raffaele Iennaco, Maria Lachgar, Marco Tullio Rigoli, Berta Marcó de la Cruz, Pietro Lo Riso, Erika Tenderini, Flavia Troglio, Marco de Simone, Isabel Liste-Noya, Stefano Piccolo, Giuseppe Macino, Massimiliano Pagani, Elena Cattaneo, Giuseppe Testa
Satb2 regulates proliferation and nuclear integrity of pre-osteoblasts
Todd Dowrey, Evelyn E Schwager, Julieann Duong, Fjodor Merkuri, Yuri A Zarate, Jennifer L Fish
Endothelial PKA targets ATG16L1 to regulate angiogenesis by limiting autophagy
Xiaocheng Zhao, Pavel Nedvetsky, Anne-Clemence Vion, Oliver Popp, Kerstin Zühlke, Gunnar Dittmar, Enno Klussmann, Holger Gerhardt
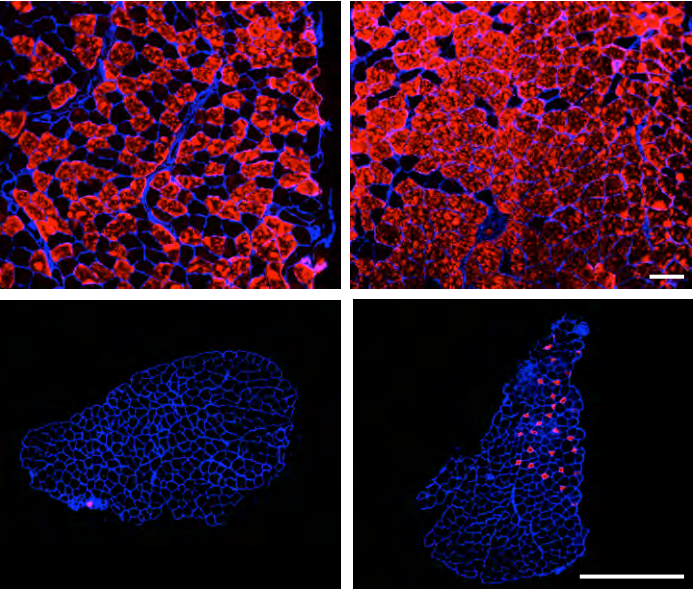
Mitogen-Activated Protein Kinase-Dependent Fiber-Type Regulation in Skeletal Muscle
Justin G. Boyer, Taejeong Song, Donghoon Lee, Xing Fu, Sakthivel Sadayappan, Jeffery D. Molkentin
A pleiotropic role for FGF signaling in mammary gland stromal fibroblasts
Zuzana Koledova, Jakub Sumbal
Aurora kinase A mediated phosphorylation of mPOU is critical for skeletal muscle differentiation
Dhanasekaran Karthigeyan, Arnab Bose, Ramachandran Boopathi, Vinay Jaya Rao, Hiroki Shima, Narendra Bharathy, Kazuhiko Igarashi, Reshma Taneja, Tapas K. Kundu
Janus effect of glucocorticoids on differentiation of muscle fibro/adipogenic progenitors
Andrea Cerquone Perpetuini, Alessio Reggio, Mauro Cerretani, Giulio Giuliani, Marisabella Santoriello, Roberta Stefanelli, Alessandro Palma, Steven Harper, Luisa Castagnoli, Alberto Bresciani, Gianni Cesareni
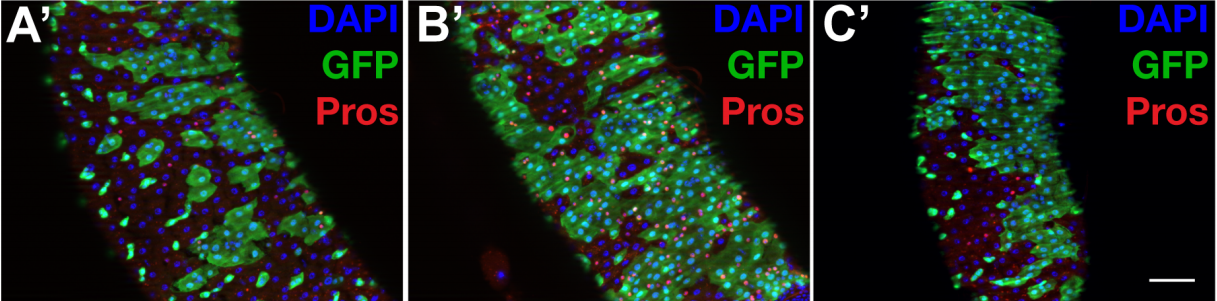
The WT1-like transcription factor Klumpfuss maintains lineage commitment in the intestine
Jerome Korzelius, Tal Ronnen-Oron, Maik Baldauf, Elke Meier, Pedro Sousa-Victor, Heinrich Jasper
Drosophila ovarian germline stem cell cytocensor projections dynamically receive and attenuate BMP signaling
Scott G. Wilcockson, Hilary L. Ashe
“ER-Ca2+ sensor STIM regulates neuropeptides required for development under nutrient restriction in Drosophila”
Megha, Christian Wegener, Gaiti Hasan
A genetic screen using the Drosophila melanogaster TRiP RNAi collection to identify metabolic enzymes required for eye development
Rose C. Pletcher, Sara L. Hardman, Sydney F. Intagliata, Rachael L. Lawson, Aumunique Page, Jason M. Tennessen
Random birth order and bursty Notch ligand expression drive the stochastic AC/VU cell fate decision in C. elegans
Simone Kienle, Nicola Gritti, Jason R. Kroll, Ana Kriselj, Yvonne Goos, Jeroen S. van Zon
Developmental trajectory of the Caenorhabditis elegans nervous system governs its structural organization
Anand Pathak, Nivedita Chatterjee, Sitabhra Sinha
Disrupting butterfly microbiomes does not affect host survival and development
Kruttika Phalnikar, Krushnamegh Kunte, Deepa Agashe
| Morphogenesis & mechanics
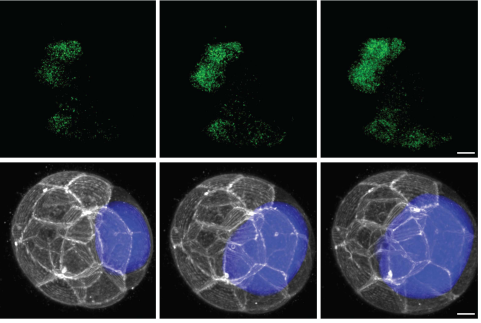
Lumen expansion facilitates epiblast-primitive endoderm fate specification in the mouse blastocyst formation.
Allyson Quinn Ryan, Chii Jou Chan, Francois Graner, Takashi Hiiragi
Septins Coordinate with Microtubules and Actin to Initiate Cell Morphogenesis
Diana Bogorodskaya, Lee A. Ligon
Rule-governed Dynamic Stochastic Equilibration of Multicellular Motion In Vivo During Olfactory Neurogenesis
Vijay Warrier, Celine Cluzeau, Bi-Chang Chen, Abigail Green-Saxena, Dani E Bergey, Eric Betzig, Ankur Saxena

Interkinetic nuclear migration in the zebra1sh retina as a diffusive process
Afnan Azizi, Anne Herrmann, Yinan Wan, Salvador J. R. P. Buse, Philipp J. Keller, Raymond E. Goldstein, William A. Harris
Kindlin-3 Mutation in Mesenchymal Stem Cells Results in Enhanced Chondrogenesis
Bethany A. Kerr, Lihong Shi, Alexander H. Jinnah, Jeffrey S. Willey, Donald P. Lennon, Arnold I. Caplan, Tatiana V. Byzova
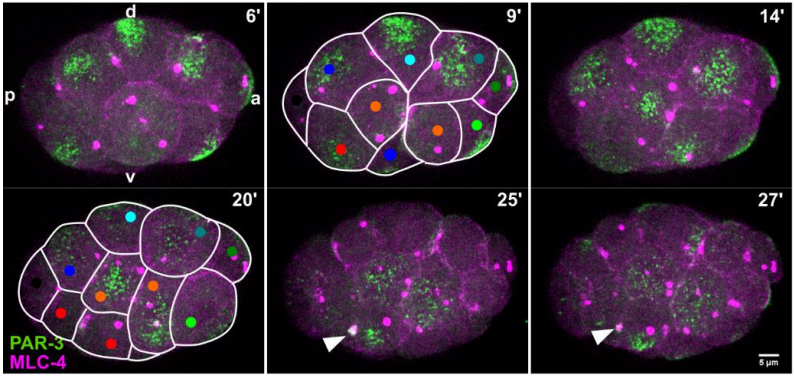
Planar cell polarity in the C. elegans embryo emerges by differential retention of aPARs at cell-cell contacts
Priyanka Dutta, Devang Odedra, Christian Pohl
α-integrins dictate distinct modes of type IV collagen recruitment to basement membranes
Ranjay Jayadev, Qiuyi Chi, Daniel P. Keeley, Eric L. Hastie, David R. Sherwood
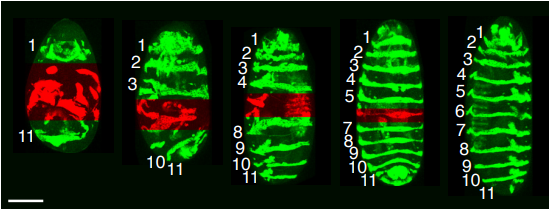
Embryonic geometry underlies phenotypic variation in decanalized conditions
A. Huang, T. E. Saunders
Apical transport of Crumbs maintains epithelial cell polarity
Mario Aguilar-Aragon, Georgina Fletcher, Barry James Thompson
Septate junctions regulate gut homeostasis through regulation of stem cell proliferation and enterocyte behavior in Drosophila
Yasushi Izumi, Kyoko Furuse, Mikio Furuse
Oriented basement membrane fibrils provide a memory for F-actin planar polarization via the Dystrophin-Dystroglycan complex during tissue elongation
Fabiana Cerqueira Campos, Hervé Alégot, Cynthia Dennis, Cornelia Fritsch, Adam Isabella, Pierre Pouchin, Olivier Bardot, Sally Horne-Badovinac, Vincent Mirouse
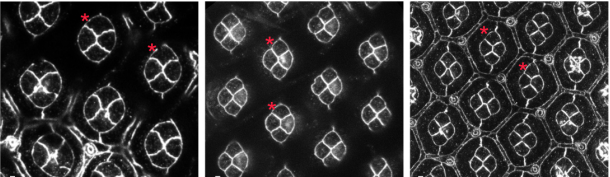
Neph/Nephrin-like adhesion and tissue level pulling forces regulate cell intercalation during Drosophila retina development.
Laura Blackie, Melda Tozluoglu, Mateusz Trylinski, Rhian Walther, Yanlan Mao, Francois Schweisguth, Franck Pichaud
Ring canals permit extensive cytoplasm sharing among germline cells independent of fusomes in Drosophila testes
Ronit S. Kaufman, Kari L. Price, Katelynn M. Mannix, Kathleen Ayers, Andrew M. Hudson, Lynn Cooley
Distinct RhoGEFs activate apical and junctional actomyosin contractility under control of G proteins during epithelial morphogenesis
Alain Garcia De Las Bayonas, Jean-Marc Philippe, Annemarie C. Lellouch, Thomas Lecuit
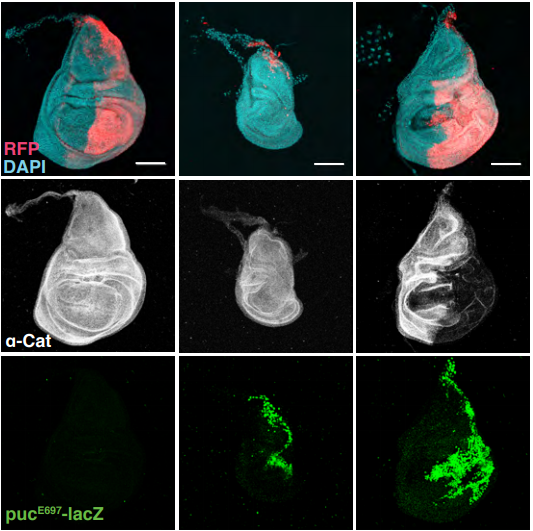
Role of α-Catenin and its mechanosensing properties in the regulation of Hippo/YAP-dependent tissue growth
Ritu Sarpal, Victoria Yan, Lidia Kazakova, Luka Sheppard, Ulrich Tepass
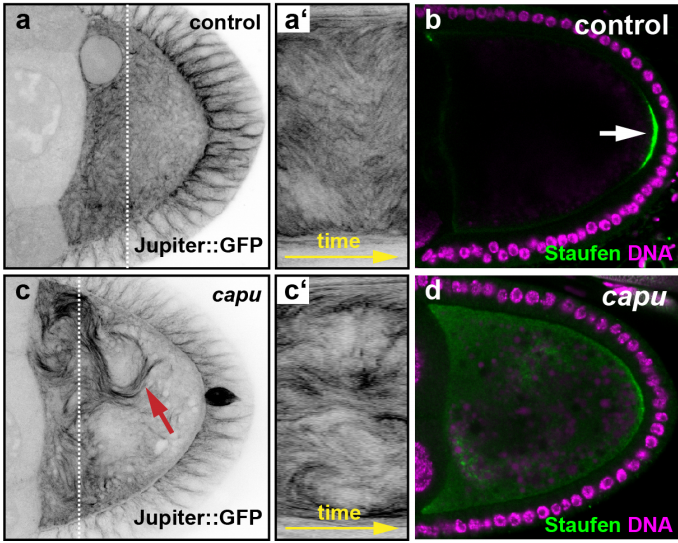
Optical flow analysis reveals that Kinesin-mediated advection impacts on the orientation of microtubules
Maik Drechsler, Lukas F. Lang, Hendrik Dirks, Martin Burger, Carola-Bibiane Schönlieb, Isabel M. Palacios
Adult Drosophila Muscle Morphometry through MicroCT reveals dynamics during aging
Dhananjay Chaturvedi, Sunil Prabhakar, Aman Aggarwal, K VijayRaghavan
Cyto-architecture constrains a photoactivation induced tubulin gradient in the syncytial Drosophila embryo
Sameer Thukral, Bivash Kaity, Bipasha Dey, Swati Sharma, Amitabha Nandi, Mithun Mitra, Richa Rikhy
A novel, dynein-independent mechanism focuses the endoplasmic reticulum around spindle poles in dividing Drosophila spermatocytes
Darya Karabasheva, Jeremy Thomas Smyth
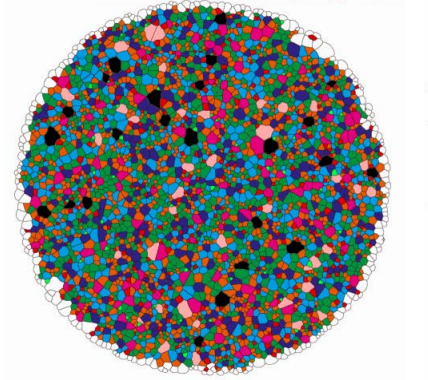
Minimisation of surface energy drives apical epithelial organisation and gives rise to Lewis’ law
Marco Kokic, Antonella Iannini, Gema Villa Fombuena, Fernando Casares, Dagmar Iber
Aboave-Weaire’s law in epithelia results from an angle constraint in contiguous polygonal lattices
Roman Vetter, Marco Kokic, Harold Gomez, Leonie Hodel, Bruno Gjeta, Antonella Iannini, Gema Villa-Fombuena, Fernando Casares, Dagmar Iber
Nanopore formation in the cuticle of an insect olfactory sensillum
Toshiya Ando, Sayaka Sekine, Sachi Inagaki, Kazuyo Misaki, Laurent Badel, Hiroyuki Moriya, Mustafa M Sami, Yuki Itakura, Takahiro Chihara, Hokto Kazama, Shigenobu Yonemura, Shigeo Hayashi
| Genes & genomes
Germ Granules Functions are Memorized by Transgenerationally Inherited Small RNAs
Itamar Lev, Itai Antoine Toker, Yael Mor, Anat Nitzan, Guy Weintraub, Ornit Bhonkar, Itay Ben Shushan, Uri Seroussi, Julie Claycomb, Hila Gingold, Ronen Zaidel-Bar, Oded Rechavi
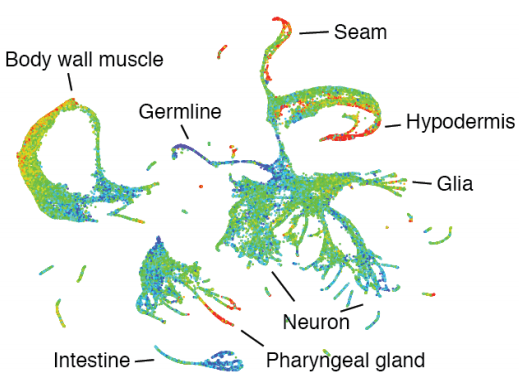
A lineage-resolved molecular atlas of C. elegans embryogenesis at single cell resolution
Jonathan S. Packer, Qin Zhu, Chau Huynh, Priya Sivaramakrishnan, Elicia Preston, Hannah Dueck, Derek Stefanik, Kai Tan, Cole Trapnell, Junhyong Kim, Robert H. Waterston, John I. Murray
HRPK-1, a conserved KH-domain protein, modulates microRNA activity during Caenorhabditis elegans development
Li Li, Isana Veksler-Lublinsky, Anna Y. Zinovyeva
The C. elegans SET-2 histone methyltransferase maintains germline fate by preventing progressive transcriptomic deregulation across generations
Valérie J. Robert, Andrew K. Knutson, Andreas Rechtsteiner, Gaël Yvert, Susan Strome, Francesca Palladino

Multi-enhancer transcriptional hubs confer phenotypic robustness
Albert Tsai, Mariana Alves, Justin Crocker
The transcription factor Odd-paired regulates temporal identity in transit-amplifying neural progenitors via an incoherent feed-forward loop
Merve Deniz Abdusselamoglu, Elif Eroglu, Thomas R Burkard, Jürgen Knoblich
The role of insulators in transgene transvection in Drosophila
Pawel Piwko, Ilektra Vitsaki, Ioannis Livadaras, Christos Delidakis
BEN-solo factors partition active chromatin to ensure proper gene activation in Drosophila
Malin Ueberschar, Huazhe Wang, Chun Zhang, Enen Guo, Eric Lai, Jiayu Wen, Qi Dai
Modular tissue-specific regulation of doublesex underpins sexually dimorphic development in Drosophila
Gavin R. Rice, Olga Barmina, David Luecke, Kevin Hu, Michelle Arbeitman, Artyom Kopp
Coordinating Receptor Expression and Wiring Specificity in Olfactory Receptor Neurons
Hongjie Li, Tongchao Li, Felix Horns, Jiefu Li, Qijing Xie, Chuanyun Xu, Bing Wu, Justus Kebschull, David Vacek, Anthony Xie, David Luginbuhl, Stephen Quake, Liqun Luo
Yunze Liu, Xiaojie Sun, Aijun Qu
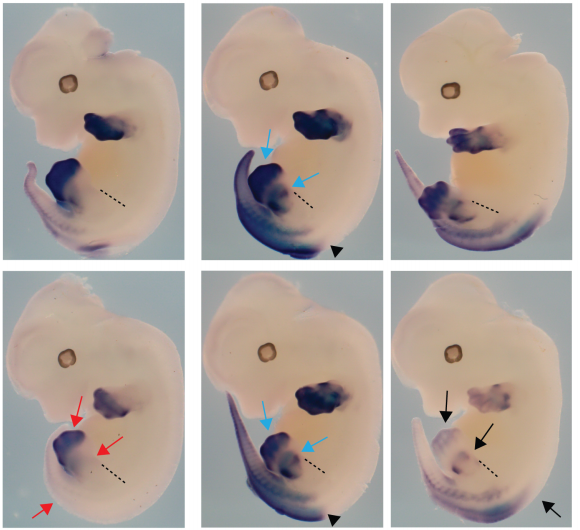
Constrained Transcriptional Polarity in the Organization of Mammalian Hox Gene Clusters
Fabrice Darbellay, Célia Bochaton, Lucille Lopez-Delisle, Bénédicte Mascrez, Patrick Tschopp, Saskia Delpretti, Jozsef Zakany, Denis Duboule
Impact of Genome Architecture Upon the Functional Activation and Repression of Hox Regulatory Landscapes
Eddie Rodríguez-Carballo, Lucille Lopez-Delisle, Nayuta Yakushiji-Kaminatsui, Asier Ullate-Agote, Denis Duboule
Pluripotency factors regulate the onset of Hox cluster activation in the early embryo
Elena Lopez-Jimenez, Julio Sainz de Aja, Claudio Badia-Careaga, Antonio Barral, Isabel Rollan, Raquel Rouco, Elisa Santos, María Tiana, Jesus Victorino, Hector Sanchez-Iranzo, Rafael D Acemel, Carlos Torroja, Javier Adan, Eduardo Andres-Leon, Jose Luis Gomez-Skarmeta, Giovanna Giovinazzo, Fatima Sanchez-Cabo, Miguel Manzanares
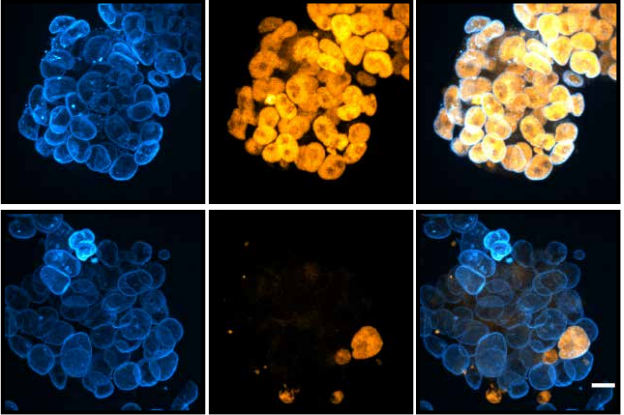
Cohesin disrupts polycomb-dependent chromosome interactions
JDP Rhodes, A Feldmann, B Hernández-Rodríguez, N Díaz, JM Brown, NA Fursova, NP Blackledge, P Prathapan, P Dobrinic, M Huseyin, A Szczurek, K Kruse, KA Nasmyth, VJ Buckle, JM Vaquerizas, RJ Klose
Three-dimensional genome reorganization during mouse spermatogenesis
Zhengyu Luo, Xiaorong Wang, Ruoyu Wang, Jian Chen, Yusheng Chen, Qianlan Xu, Jun Cao, Xiaowen Gong, Ji Wu, Yungui Yang, Wenbo Li, Chunsheng Han, Fei Sun, Xiaoyuan Song
Functional dissection of TADs reveals non-essential and instructive roles in regulating gene expression
Alexandra Despang, Robert Schöpflin, Martin Franke, Salaheddine Ali, Ivana Jerkovic, Christina Paliou, Wing-Lee Chan, Bernd Timmermann, Lars Wittler, Martin Vingron, Stefan Mundlos, Daniel M. Ibrahim
DUX is a non-essential synchronizer of zygotic genome activation
Alberto De Iaco, Sonia Verp, Sandra Offner, Didier Trono
Neuronal histone methyltransferase EZH2 regulates neuronal morphogenesis, synaptic plasticity, and cognitive behavior of mice
Mei Zhang, Yong Zhang, Qian Xu, Joshua Crawford, Cheng Qian, Guo-Hua Wang, Eastman Lewis, Philip Hall, Gül Dolen, Richard L. Huganir, Jiang Qian, Xin-Zhong Dong, Mikhail V. Pletnikov, Chang-Mei Liu, Feng-Quan Zhou
Transcriptional maintenance of cortical somatostatin interneuron subtype identity during migration
Hermany Munguba, Kasra Nikouei, Hannah Hochgerner, Polina Oberst, Alexandra Kouznetsova, Jesper Ryge, Renata Bastista-Brito, Ana Belen Munoz-Manchado, Jennie Close, Sten Linnarsson, Jens Hjerling Leffler
Systematic Comparison of High-throughput Single-Cell and Single-Nucleus Transcriptomes during Cardiomyocyte Differentiation
Alan Selewa, Ryan Dohn, Heather Eckart, Stephanie Lozano, Bingqing Xie, Eric Gauchat, Reem Elorbany, Katherine Rhodes, Jonathan Burnett, Yoav Gilad, Sebastian Pott, Anindita Basu
Whole-genome and RNA sequencing reveal variation and transcriptomic coordination in the developing human prefrontal cortex
Donna M. Werling, Sirisha Pochareddy, Jinmyung Choi, Joon-Yong An, Brooke Sheppard, Minshi Peng, Zhen Li, Claudia Dastmalchi, Gabriel Santpere, Andre M. M. Sousa, Andrew T. N. Tebbenkamp, Navjot Kaur, Forrest O. Gulden, Michael S. Breen, Lindsay Liang, Michael C. Gilson, Xuefang Zhao, Shan Dong, Lambertus Klei, A. Ercument Cicek, Joseph D. Buxbaum, Homa Adle-Biassette, Jean-Leon Thomas, Kimberly A. Aldinger, Diana R. O’Day, Ian A. Glass, Noah A. Zaitlen, Michael E. Talkowski, Kathryn Roeder, Matthew W. State, Bernie Devlin, Stephan J. Sanders, Nenad Sestan
Single-Cell RNA Sequencing Reveals Regulatory Mechanism for Trophoblast Cell-Fate Divergence in Human Peri-Implantation Embryo
Bo Lv, Qin An, Qiao Zeng, Ping Lu, Xianmin Zhu, Yazhong Ji, Guoping Fan, Zhigang Xue
Unique Trophoblast Chromatin Environment Mediated by the PcG Protein SFMBT2
Priscilla Tang, Kamelia Miri, Susannah Varmuza
Circular RNA profiling in the oocyte and cumulus cells reveals that circARMC4 is essential for porcine oocyte maturation
Zubing Cao, Di Gao, Tengteng Xu, Ling Zhang, Xu Tong, Dandan Zhang, Yiqing Wang, Wei Ning, Xin Qi, Yangyang Ma, Kaiyuan Ji, Tong Yu, Yunsheng Li, Yunhai Zhang
p63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes
Jieqiong Qu, Guoqiang Yi, Huiqing Zhou
Single-cell transcriptomics reveals multiple neuronal cell types in human midbrain-specific organoids
Lisa M Smits, Stefano Magni, Kamil Grzyb, Paul MA Antony, Rejko Krueger, Alexander Skupin, Silvia Bolognin, Jens Schwamborn
A pivotal genetic program controlled by thyroid hormone during the maturation of GABAergic neurons in mice
Sabine Richard, Romain Guyot, Martin Rey-Millet, Margaux Prieux, Suzy Markossian, Denise Aubert, Frederic Flamant
PRDM16 establishes lineage-specific transcriptional program to promote temporal progression of neural progenitors in the mouse neocortex
Li He, Jennifer Jones, Weiguo He, Bryan Bjork, Jiayu Wen, Qi Dai
Enhancer-promoter association determines Sox2 transcription regulation in mouse pluripotent cells
Lei Huang, Qing Li, Qitong Huang, Siyuan Kong, Xiusheng Zhu, Yanling Peng, Yubo Zhang
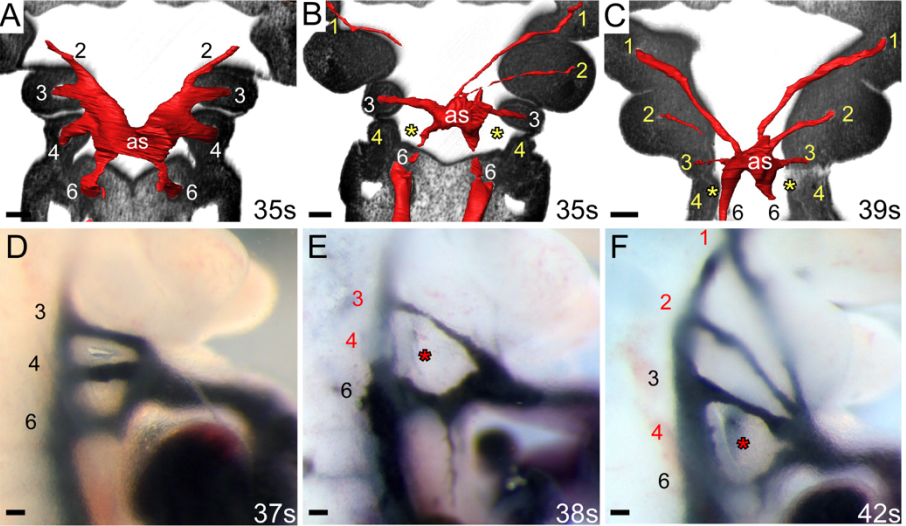
Pax9 is required for cardiovascular development and interacts with Tbx1 in the pharyngeal endoderm to control 4th pharyngeal arch artery morphogenesis.
Helen M Phillips, Catherine A Stothard, Wasay Mohiuddin Shaikh Qureshi, Anastasia I Kousa, Alberto Briones-Leon, Ramada Khasawneh, Rachel Sanders, Silvia Mazotta, Rebecca Dodds, Kerstin Seidel, Timothy Bates, Mitsushiro Nakatomi, Simon Cockell, Jurgen E Schneider, Timothy J Mohun, Rene Maehr, Ralf Kist, Heiko Peters, Simon D Bamforth
The Pax6 master control gene initiates spontaneous retinal development via a self-organising Turing network
Timothy Grocott, Estefania Lozano-Velasco, Gi Fay Mok, Andrea E Münsterberg
Identification of master regulators in goblet cells and Paneth cells using transcriptomics profiling of gut organoids and multi-layered networks
Agatha Treveil, Padhmanand Sudhakar, Zoe J Matthews, Tomasz Wrzesinski, Emily J Jones, Marton Olbei, Isabelle Hautefort, Lindsay J Hall, Simon R Carding, Ulrike Mayer, Penny P Powell, Tom Wileman, Federica Di Palma, Wilfried Haerty, Tamas Korcsmaros
Characterizing the nuclear and cytoplasmic transcriptomes in developing and mature human cortex uncovers new insight into psychiatric disease gene regulation
Amanda J. Price, Taeyoung Hwang, Ran Tao, Emily E. Burke, Anandita Rajpurohi, Joo Heon Shin, Thomas M. Hyde, Joel E. Kleinman, Andrew E. Jaffe, Daniel R. Weinberger
| Stem cells, regeneration & disease modelling
Cultured pluripotent planarian stem cells retain potency and express proteins from exogenously introduced mRNAs
Kai Lei, Sean A McKinney, Eric J Ross, Heng-Chi Lee, Alejandro Sanchez Alvarado
Human cortical neural stem cells generate regional organizer states in vitro before committing to excitatory neuronal fates
Nicola Micali, Suel-Kee Kim, Marcelo Diaz-Bustamante, Genevieve Stein-O’Brien, Seungmae Seo, Joo-Heon Shin, Brian G. Rash, Shaojie Ma, Nicolas A. Olivares, Jon Arellano, Kristen R. Maynard, Elana J. Fertig, Alan J. Cross, Roland Burli, Nicholas J. Brandon, Daniel R. Weinberger, Joshua G. Chenoweth, Daniel J. Hoeppner, Nenad Sestan, Pasko Rakic, Carlo Colantuoni, Ronald D. McKay
Identification of a human adult cardiac stem cell population with neural crest origin
Anna Höving, Madlen Merten, Kazuko Elena Schmidt, Isabel Faust, Lucia Mercedes Ruiz-Perera, Henning Hachmeister, Sebastian-Patrick Sommer, Buntaro Fujita, Thomas Pühler, Thomas Huser, Johannes Greiner, Barbara Kaltschmidt, Jan Gummert, Cornelius Knabbe, Christian Kaltschmidt
Embryonic signals perpetuate polar-like trophoblast stem cells and pattern the blastocyst axis
Javier Frias-Aldeguer, Maarten Kip, Judith Vivié, Linfeng Li, Anna Alemany, Jeroen Korving, Frank Darmis, Alexander van Oudenaarden, Clemens A. Van Blitterswijk, Niels Geijsen, Nicolas C. Rivron
Genes essential for embryonic stem cells are associated with neurodevelopmental disorders
Shahar Shohat, Sagiv Shifman
Quantitative multiplexed ChIP reveals global alterations that shape promoter bivalency in ground state embryonic stem cells
Banushree Kumar, Simon J Elsässer
Depletion of resident muscle stem cells inhibits muscle fiber hypertrophy induced by lifelong physical activity
Davis A. Englund, Kevin A. Murach, Cory M. Dungan, Vandré C. Figueiredo, Ivan J. Vechetti Jr., Esther E. Dupont-Versteegden, John J. McCarthy, Charlotte A. Peterson
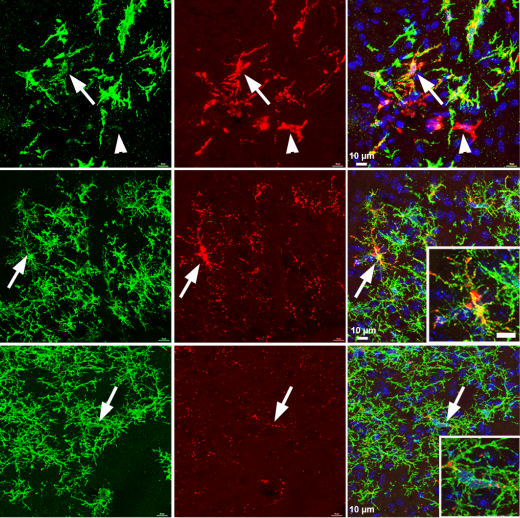
Xenotransplantation of Human PSC-derived Microglia Creates a Chimeric Mouse Brain Model that Recapitulates Features of Adult Human Microglia
Ranjie Xu, Andrew Boreland, Anthony Posyton, Kelvin Kwan, Ronald P. Hart, Peng Jiang
The Gag Protein PEG10 Binds to RNA and Regulates Trophoblast Stem Cell Lineage Specification
Mona Abed, Erik Verschueren, Hanna Budayeva, Peter Liu, Donald S. Kirkpatrick, Rohit Reja, Sarah K. Kummerfeld, Joshua D. Webster, Sarah Gierke, Mike Reichelt, Keith R. Anderson, Robert J Newman, Merone Roose-Girma, Zora Modrusan, Hazal Pektas, Emin Maltepe, Kim Newton, Vishva M. Dixit
Simultaneous tracking of division and differentiation from individual hematopoietic stem and progenitor cells reveals within-family homogeneity despite population heterogeneity
Tamar Tak, Giulio Prevedello, Gaël Simon, Noémie Paillon, Ken R. Duffy, Leïla Perié
The evolutionary dynamics and fitness landscape of clonal haematopoiesis
Caroline J. Watson, Alana Papula, Yeuk P. G. Poon, Wing H. Wong, Andrew L. Young, Todd E. Druley, Daniel S. Fisher, Jamie R. Blundell
Ex vivo expansion of skeletal muscle stem cells with a novel small compound inhibitor of eIF2α dephosphorylation
Graham Lean, Matt Halloran, Oceane Mariscal, Solene Jamet, Jean-Phillip Lumb, Colin Crist
Non-proliferative adult neurogenesis in neural crest-derived stem cells isolated from human periodontal ligament
Carlos Bueno, Marta Martínez-Morga, Salvador Martínez

Generic and context-dependent gene modulations during Hydra whole body regeneration
Yvan Wenger, Wanda Buzgariu, Chrystelle Perruchoud, Gregory Loichot, Brigitte Galliot
Mouth Function Determines The Shape Oscillation Pattern In Regenerating Hydra Tissue Spheres
R. Wang, T. Goel, K. Khazoyan, Z. Sabry, H.J. Quan, P.H. Diamond, E.M.S. Collins
Single cell multi-omics analysis reveals novel roles for DNA methylation in sensory neuron injury responses
Youjin Hu, Qin An, Guoping Fan
Proteomics analysis of extracellular matrix remodeling during zebrafish heart regeneration
Anna Garcia-Puig, Jose Luis Mosquera, Senda Jiménez-Delgado, Cristina García-Pastor, Ignasi Jorba, Daniel Navajas, Francesc Canals, Angel Raya
A smooth muscle-like niche facilitates lung epithelial regeneration
Alena Moiseenko, Ana Ivonne Vazquez-Armendariz, Xuran Chu, Stefan Guenther, Kevin Lebrigand, Vahid Kheirollahi, Susanne Herold, Thomas Braun, Bernard Mari, Stijn De Langhe, Chengshui Chen, Xiaokun Li, Werner Seeger, Jin-San Zhang, Saverio Bellusci, Elie El Agha
The gut microbiota regulates mouse biliary regenerative responses
Wenli Liu, Chao Yan, Bo Zhang, Renjin Chen, Qian Yu, Xiangyang Li, Yuzhao Zhang, Hui Hua, Yanxia Wei, Yanbo Kou, Zhuanzhuan Liu, Renxian Tang, Kuiyang Zheng, Yugang Wang
Bacteria are required for regeneration of the Xenopus tadpole tail
Thomas F. Bishop, Caroline W. Beck
Monosodium Iodoacetate delays regeneration and inhibits hypertrophy in skeletal muscle cells in vitro
Rowan P Rimington, Darren J Player, Neil R.W Martin, Mark P Lewis
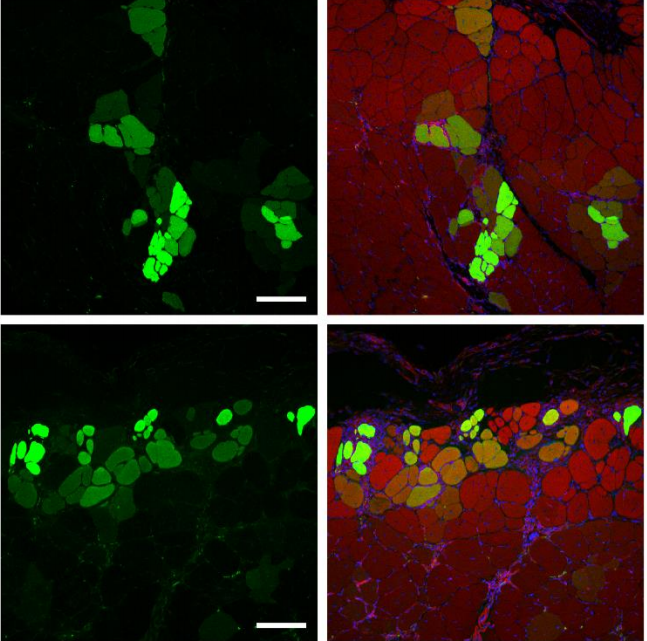
Wnt7a-releasing synthetic hydrogel enhances local skeletal muscle regeneration and muscle stem cell engraftment
Woojin M. Han, Mahir Mohiuddin, Shannon E. Anderson, Andrés J. García, Young C. Jang
Effects of estrogen on Survival and Neuronal Differentiation of adult human olfactory bulb neural stem Cells Transplanted into Spinal Cord Injured Rats
S Rezk, A Althani, A Abd-Elmaksoud, M Kassab, A Farag, S Lashen, C Cenciarelli, T Caceci, HE Marei
Characterisation of l(3)tb as a novel tumour suppressor allele of DCP2 in Drosophila melanogaster
Rakesh Mishra, Rohit Kunar, LOLITIKA MANDAL, Debasmita Pankaj Alone, Shanti Chandrasekharan, Anand Krishna Tiwari, Ashim Mukherjee, Madhu Gwaldas Tapadia, Jagat Kumar Roy
Missense Mutations in the Human Nanophthalmos Gene TMEM98 Cause Retinal Defects in the Mouse
Sally H. Cross, Lisa Mckie, Margaret Keighren, Katrine West, Caroline Thaung, Tracey Davey, Dinesh C. Soares, Luis Sanchez-Pulido, Ian J. Jackson
Human liver organoids; a patient-derived primary model for HBV Infection and Related Hepatocellular Carcinoma
Elisa De Crignis, Fabrizia Carofiglio, Panagiotis Moulos, Monique M.A. Verstegen, Shahla Romal, Mir Mubashir Khalid, Farzin Pourfarzad, Christina Koutsothanassis, Helmuth Gehart, Tsung Wai Kan, Robert-Jan Palstra, Charles Boucher, Jan M.N. Ijzermans, Meritxell Huch, Sylvia F. Boj, Robert Vries, Hans Clevers, Luc van der Laan, Pantelis Hatzis, Tokameh Mahmoudi
A segregating human allele of SPO11 modeled in mice disrupts timing and amounts of meiotic recombination, causing oligospermia and a decreased ovarian reserve.
Tina N Tran, John C. Schimenti
Intraventricular CXCL12 is neuroprotective and increases neurogenesis in a murine model of stroke
Laura N. Zamproni, Marimelia A. Porcionatto
Reversing Abnormal Neural Development by Inhibiting OLIG2 in Down Syndrome Human iPSC Brain Organoids and Neuronal Mouse Chimeras
Ranjie Xu, Andrew T Brawner, Shenglan Li, JingJing Liu, Hyosung Kim, Haipeng Xue, Zhiping P. Pang, Woo-Yang Kim, Ronald P. Hart, Ying Liu, Peng Jiang
Modeling motor neuron resilience in ALS using stem cells
Ilary Allodi, Jik Nijssen, Julio Aguila Benitez, Christoph Schweingruber, Andrea Fuchs, Gillian Bonvicini, Ming Cao, Ole Kiehn, Eva Hedlund
Transcriptomic analysis of the BDNF-induced JAK/STAT pathway in neurons: a window into epilepsy-associated gene expression
Kathryn M Hixson, Meaghan C Cogswell, Amy R Brooks-Kayal, Shelley J Russek
Glioblastomas derived from genetically modified pluripotent stem cells recapitulate pathobiology
Tomoyuki Koga, Jorge A Benitez, Isaac A Chaim, Sebastian Markmiller, Alison D Parisian, Kristen M Turner, Florian M Hessenauer, Matteo D’Antonio, Nam-phuong D Nguyen, Shahram Saberi, Jianhui Ma, Shunichiro Miki, Antonia D Boyer, John Ravits, Kelly A Frazer, Vineet Bafna, Clark C Chen, Paul S Mischel, Gene W Yeo, Frank B Furnari
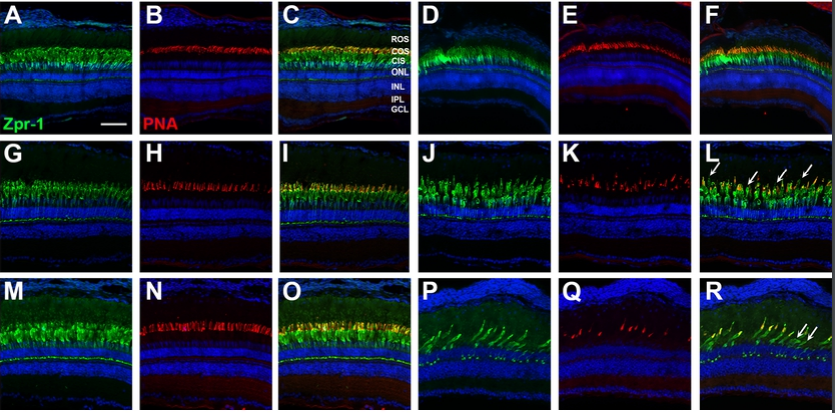
Ciliary Genes arl13b, ahi1 and cc2d2a Differentially Modify Expression of Visual Acuity Phenotypes but do not Enhance Retinal Degeneration due to Mutation of cep290 in Zebrafish
Emma M. Lessieur, Ping Song, Gabrielle C. Nivar, Ellen M. Piccillo, Joseph Fogerty, Richard Rozic, Brian D. Perkins
| Plant development
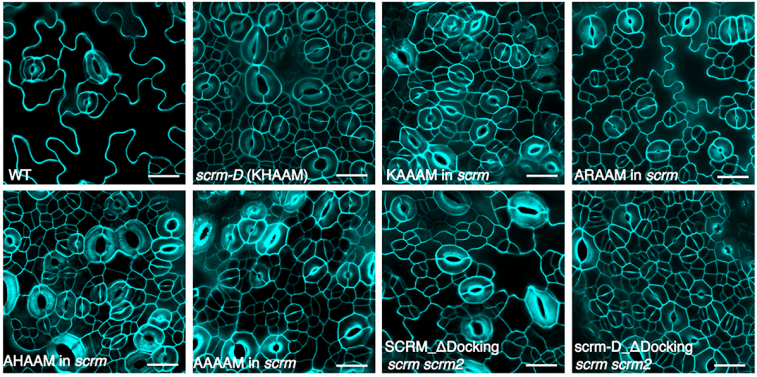
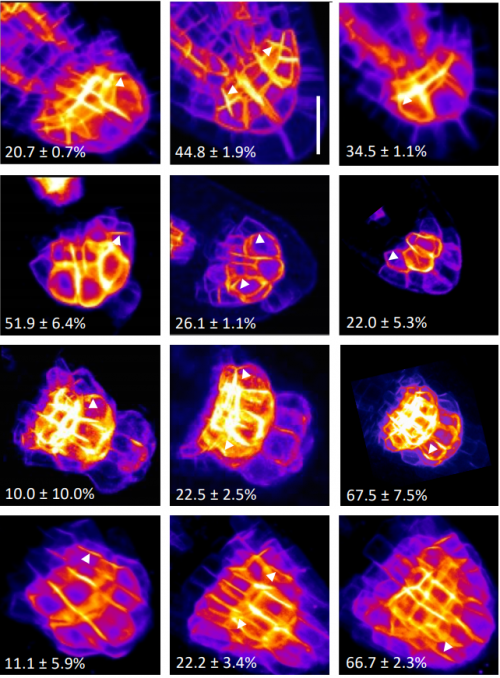
Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP Kinases to SPEECHLESS
Aarthi Putarjunan, Jim Ruble, Ashutosh Srivastava, Chunzhao Zhao, Amanda L. Rychel, Alex K. Hofstetter, Xiaobo Tang, Jian-Kang Zhu, Florence Tama, Ning Zheng, Keiko U. Torii
Cytoskeleton members, MVBs and the ESCRT-III HvSNF7s are putative key players for protein sorting into protein bodies during barley endosperm development
Valentin Roustan, Julia Hilscher, Marieluise Weidinger, Siegfried Reipert, Azita Shabrangy, Claudia Gebert, Bianca Dietrich, Georgi Dermendjiev, Pierre-Jean Roustan, Eva Stoeger, Verena Ibl
A Musashi-Related Protein is Essential for Gametogenesis in Arabidopsis
Laura A. Moody, Ester Rabbinowitsch, Hugh G. Dickinson, Roxaana Clayton, David M. Emms, Jane A. Langdale
Epistatic Transcription Factor Networks Differentially Modulate Arabidopsis Growth and Defense
Baohua Li, Michelle Tang, Céline Caseys, Ayla Nelson, Marium Zhou, Xue Zhou, Siobhan M. Brady, Daniel J. Kliebenstein
Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots
Sascha Waidmann, Michel Ruiz Rosquete, Maria Schöller, Heike Lindner, Therese LaRue, Elizabeth Sarkel, Ivan Petřík, Kai Dünser, Shanice Martopawiro, Rashmi Sasidharan, Ondrej Novak, Krzysztof Wabnik, José R. Dinneny, Jürgen Kleine-Vehn
Two ecotype-related long non-coding RNAs in the environmental control of root growth
Thomas Blein, Coline Balzergue, Thomas Roulé, Marc Gabriel, Laetitia Scalisi, Céline Sorin, Aurélie Christ, Etienne Delannoy, Marie-Laure Martin-Magniette, Laurent Nussaume, Caroline Hartmann, Daniel Gautheret, Thierry Desnos, Martin Crespi
Metabolic labeling of RNAs uncovers hidden features and dynamics of the Arabidopsis thaliana transcriptome
Emese Xochitl Szabo, Philipp Reichert, Marie-Kristin Lehniger, Marilena Ohmer, Marcella de Francisco Amorim, Udo Gowik, Christian Schmitz-Linneweber, Sascha Laubinger
Mechanical stress initiates and sustains the morphogenesis of wavy leaf epidermal cells
Amir J Bidhendi, Bara Altartouri, Frédérick P. Gosselin, Anja Geitmann
Strain- or Stress-sensing in mechanochemical patterning by the phytohormone auxin
Jean-Daniel Julien, Alain Pumir, Arezki Boudaoud
Antagonistic and auxin-dependent phosphoregulation of columella PIN proteins controls lateral root gravitropic setpoint angle in Arabidopsis
Suruchi Roychoudhry, Katelyn Sageman-Furnas, Chris Wolverton, Heather L. Goodman, Peter Grones, Jack Mullen, Roger Hangarter, Jiri Friml, Stefan Kepinski
Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in histone H3 Lys-4 trimethylation independently from histone H2B monoubiquitination
Anne-Sophie Fiorucci, Clara Bourbousse, Lorenzo Concia, Martin Rougée, Anne-Flore Deton-Cabanillas, Gérald Zabulon, Elodie Layat, David Latrasse, SoonKap Kim, Nicole Chaumont, Bérangère Lombard, David Stroebel, Sophie Lemoine, Ammara Mohammad, Corinne Blugeon, Damarys Loew, Christophe Bailly, Chris Bowler, Moussa Benhamed, Fredy Barneche
Molecular mechanisms of Evening Complex activity in Arabidopsis
Catarina S. Silva, Aditya Nayak, Xuelei Lai, Veronique Hugouvieux, Jae-Hoon Jung, Agnès Jourdain, Irene López-Vidriero, Jose Manuel Franco-Zorrilla, François Parcy, Kishore Panigrahi, Philip A. Wigge, Max Nanao, Chloe Zubieta
Timing seed germination under changing salinity: a key role of the ERECTA receptor-kinases
Amrit Kaur Nanda, Abdeljalil El Habti, Charles Hocart, Josette Masle
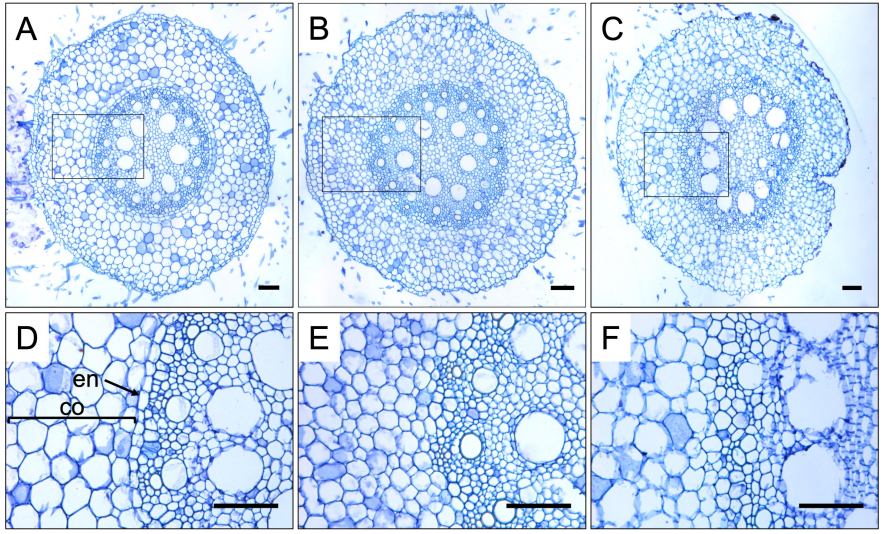
Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize
Thomas E. Hughes, Olga V. Sedelnikova, Hao Wu, Philip W. Becraft, Jane A. Langdale
Comparative transcriptomics reveals the difference in early endosperm development between maize with different amylose contents
Zhou Jian Qu, Tu Shu Xu, Kang Xiao Tian, Ting Li, Cheng Li Wang, Yue Yu Zhong, Quan Ji Xue, Wei Dong Guo
Genetic dissection of cell wall defects and the strigolactone pathway in Arabidopsis
Vicente Ramírez, Markus Pauly
Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels
M. Salehin, B. Li, M. Tang, E. Katz, L. Song, J. R. Ecker, D. Kliebenstein, M. Estelle
Low temperature triggers genome-wide hypermethylation of transposable elements and centromeres in maize
Zeineb Achour, Johann Joets, Martine Leguilloux, Hélène Sellier, Jean-Philippe Pichon, Magalie Leveugle, Hervé Duborjal, José Caius, Véronique Brunaud, Christine Paysant-Le Roux, Tristan Mary-Huard, Catherine Giauffret, Clémentine Vitte
Small RNA expression pattern in multiply inbred lines and their hybrids of maize embryo
Yong-Xin Liu
From bud formation to flowering: transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy
Noémie Vimont, Mathieu Fouché, José Antonio Campoy, Meixuezi Tong, Mustapha Arkoun, Jean-Claude Yvin, Philip A. Wigge, Elisabeth Dirlewanger, Sandra Cortijo, Bénédicte Wenden
Affordable and robust phenotyping framework to analyse root system architecture of soil-grown plants
Thibaut Bontpart, Cristobal Concha, Valerio Giuffrida, Ingrid Robertson, Kassahun Admkie, Tulu Degefu, Nigusie Girma, Kassahun Tesfaye, Teklehaimanot Haileselassie, Asnake Fikre, Masresha Fetene, Sotirios A. Tsaftaris, Peter Doerner
Tissue-specific Hi-C analyses of rice, foxtail millet and maize suggest non-canonical function of plant chromatin domains
Pengfei Dong, Xiaoyu Tu, Haoxuan Li, Jianhua Zhang, Donald Grierson, Pinghua Li, Silin Zhong
Combined morphological and transcriptomic analyses reveal genetic regulation underlying the species-specific bulbil outgrowth in Dioscorea alata L
Zhi-Gang Wu, Wu Jiang, Zheng-Ming Tao, Xiu-Zhu Guo, Xiao-Jun Pan, Wen-Hui Yu
Evo-devo & evo
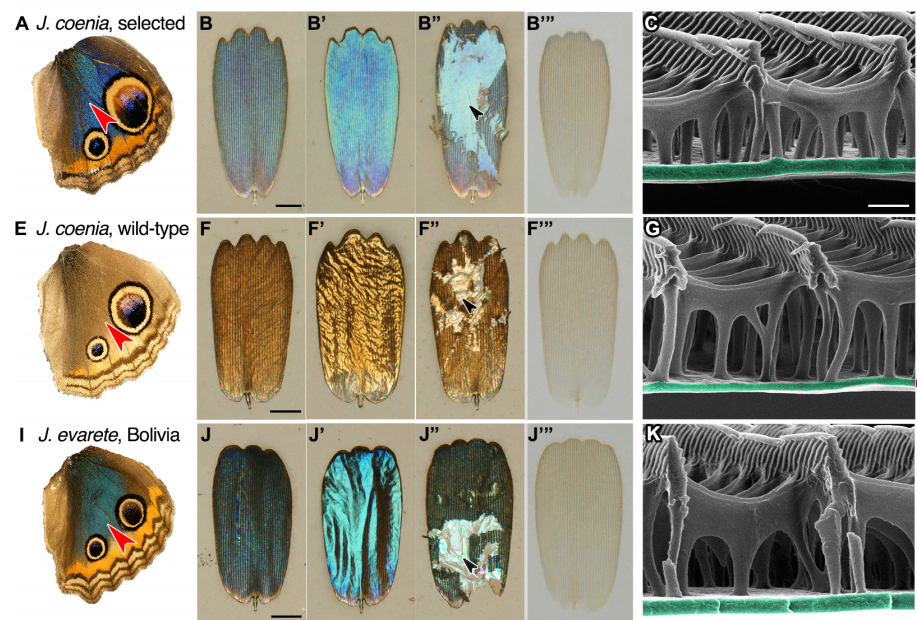
Structural color in Junonia butterflies evolves by tuning scale lamina thickness
Rachel C. Thayer, Frances I. Allen, Nipam H. Patel
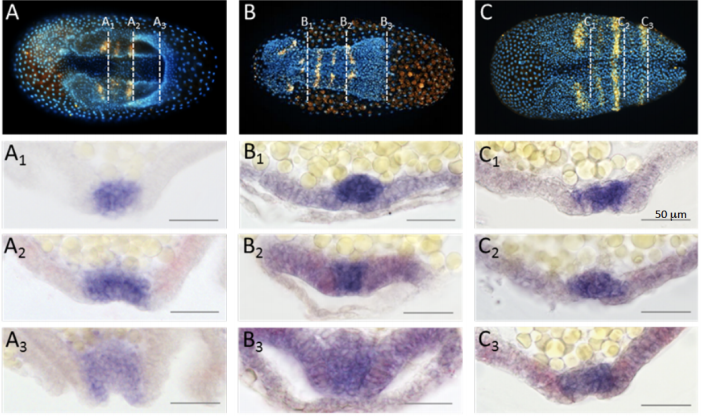
Fog signaling has diverse roles in epithelial morphogenesis in insects
Nadine Frey, Matthew A. Benton, Rodrigo Nunes da Fonseca, Cornelia von Levetzow, Dominik Stappert, Muhammad Salim Hakeemi, Kai Conrads, Matthias Pechmann, Kristen A Panfilio, Jeremy A Lynch, Siegfried Roth
Transcriptomics supports that pleuropodia of insect embryos function in degradation of the serosal cuticle to enable hatching
Barbora Konopová, Elisa Buchberger, Alastair Crisp
Hemimetabolous insects elucidate the origin of sexual development via alternative splicing
Judith Wexler, Emily K. Delaney, Xavier Belles, Coby Schal, Ayako Wada-Katsumata, Matthew Amicucci, Artyom Kopp
A cis-regulatory change underlying the motor neuron-specific loss of terminal selector gene expression in immotile tunicate larvae
Elijah K. Lowe, Claudia Racioppi, Nadine Peyriéras, Filomena Ristoratore, Lionel Christiaen, Billie J. Swalla, Alberto Stolfi
The Developmental Transcriptome for Lytechinus variegatus Exhibits Temporally Punctuated Gene Expression Changes
John D. Hogan, Jessica L. Keenan, Lingqi Luo, Dakota Y. Hawkins, Jonas Ibn-Salem, Arjun Lamba, Daphne Schatzberg, Michael L. Piacentino, Daniel T. Zuch, Amanda B. Core, Carolyn Blumberg, Bernd Timmermann, José Horacio Grau, Emily Speranza, Miguel A. Andrade-Narravo, Naoki Irie, Albert J. Poustka, Cynthia A. Bradham
Developmental transcriptomes of the sea star, Patiria miniata, illuminate the relationship between conservation of gene expression and morphological conservation
Tsvia Gildor, Gregory Cary, Maya Lalzar, Veronica Hinman, Smadar Ben-Tabou de-Leon
New non-bilaterian transcriptomes provide novel insights into the evolution of coral skeletomes
Nicola Conci, Gert Woerheide, Sergio Vargas

CRISPR-Cas9 Gene Editing in Lizards Through Microinjection of Unfertilized Oocytes
Ashley M. Rasys, Sungdae Park, Rebecca E. Ball, Aaron J. Alcala, James D. Lauderdale, Douglas B. Menke
Post-transcriptional mechanisms distinguish human and chimp forebrain progenitor cells
Daniela A Grassi, Per Ludvik Brattås, Jeovanis G Valdés, Melinda Rezeli, Marie E Jönsson, Sara Nolbrant, Malin Parmar, György Marko-Varga, Johan Jakobsson
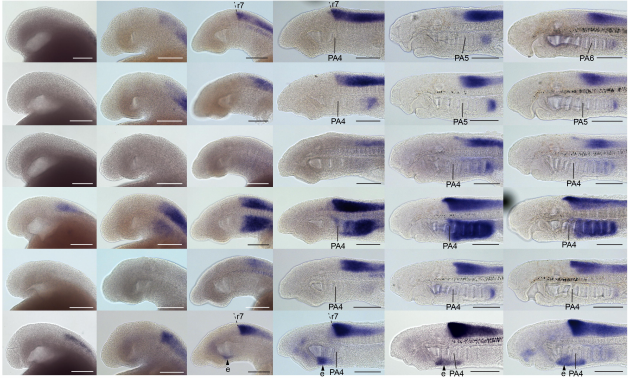
An atlas of anterior hox gene expression in the embryonic sea lamprey head: hox-code evolution in vertebrates
Hugo J. Parker, Marianne E. Bronner, Robb Krumlauf
The spindle assembly checkpoint functions during early development in non-chordate embryos
Janet Chenevert, Marianne Roca, Lydia Besnardeau, Antonella Ruggiero, Dalileh Nabi, Alex McDougall, Richard R. Copley, Elisabeth Christians, Stefania Castagnetti
A beneficial genomic rearrangement creates multiple versions of calcipressin in C. elegans
Yuehui Zhao, Jason Wan, Shweta Biliya, Shannon Brady, Daehan Lee, Erik Andersen, Fredrik Vannberg, Hang Lu, Patrick McGrath
Molecular evolution across developmental time reveals rapid divergence in early embryogenesis
Asher D. Cutter, Rose H. Garrett, Stephanie Mark, Wei Wang, Lei Sun
Evolution, not transgenerational plasticity, explains the divergence of acorn ant thermal tolerance across an urban-rural temperature cline
Ryan A. Martin, Lacy D. Chick, Aaron R. Yilmaz, Sarah E. Diamond
Genomics of expanded avian sex chromosomes shows that certain chromosomes are predisposed towards sex-linkage in vertebrates
Hanna Sigeman, Suvi Ponnikas, Pallavi Chauhan, Elisa Dierickx, M. de L. Brooke, Bengt Hansson
Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers
Iulia Darolti, Alison E. Wright, Benjamin A. Sandkam, Jake Morris, Natasha I. Bloch, Marta Farré, Rebecca C. Fuller, Godfrey R. Bourne, Denis M. Larkin, Felix Breden, Judith E. Mank
Gene-level quantitative trait mapping in an expanded C. elegans multiparent experimental evolution panel
Luke M. Noble, Matthew V. Rockman, Henrique Teotónio
Widespread gene duplication and adaptive evolution in the RNA interference pathways of the Drosophila obscura group
Danang Crysnanto, Darren Obbard
Is evolution predictable? Quantitative genetics under complex genotype-phenotype maps
Lisandro Milocco, Isaac Salazar-Ciudad
Pluripotency and the origin of animal multicellularity
Shunsuke Sogabe, William Hatleberg, Kevin Kocot, Tahsha Say, Daniel Stoupin, Kathrein Roper, Selene Fernandez-Valverde, Sandie Degnan, Bernard Degnan
A chronology of multicellularity evolution in cyanobacteria
Katrin Hammerschmidt, Giddy Landan, Fernando Domingues Kümmel Tria, Tal Dagan
The quail as an avian model system: its genome provides insights into social behaviour, seasonal biology and infectious disease response
Katrina Morris, Matthew M Hindle, Simon Boitard, David W Burt, Angela F Danner, Lel Eory, Heather L Forrest, David Gourichon, Jerome Gros, LaDeana Hillier, Thierry Jaffredo, Hanane Khoury, Rusty Lansford, Christine Leterrier, Andrew Loudon, Andrew S Mason, Simone L Meddle, Francis Minvielle, Patrick Minx, Frederique Pitel, J Patrick Seiler, Tsuyoshi Shimmura, Chad Tomlinson, Alain Vignal, Robert G Webster, Takashi Yoshimura, Wesley C Warren, Jacqueline Smith
The genome of the blind soil-dwelling and ancestrally wingless dipluran Campodea augens, a key reference hexapod for studying the emergence of insect innovations
Mosè Manni, Felipe A. Simao, Hugh M. Robertson, Marco A. Gabaglio, Robert M. Waterhouse, Bernhard Misof, Oliver Niehuis, Nikolaus Szucsich, Evgeny M. Zdobnov

Hybrid genome assembly and annotation of Danionella translucida
Mykola Kadobianskyi, Lisanne Schulze, Markus Schuelke, Benjamin Judkewitz
A draft genome sequence of the miniature parasitoid wasp, Megaphragma amalphitanum
Artem V. Nedoluzhko, Fedor S. Sharko, Brandon M. Lê, Svetlana V. Tsygankova, Eugenia S. Boulygina, Sergey M. Rastorguev, Alexey S. Sokolov, Fernando Rodriguez, Alexander M. Mazur, Alexey A. Polilov, Richard Benton, Michael B. Evgen’ev, Irina R. Arkhipova, Egor B. Prokhortchouk, Konstantin G. Skryabin
Cell biology
Oncogenic signaling alters cell shape and mechanics to facilitate cell division under confinement
Helen K. Matthews, Sushila Ganguli, Katarzyna Plak, Anna V. Taubenberger, Matthieu Piel, Jochen Guck, Buzz Baum
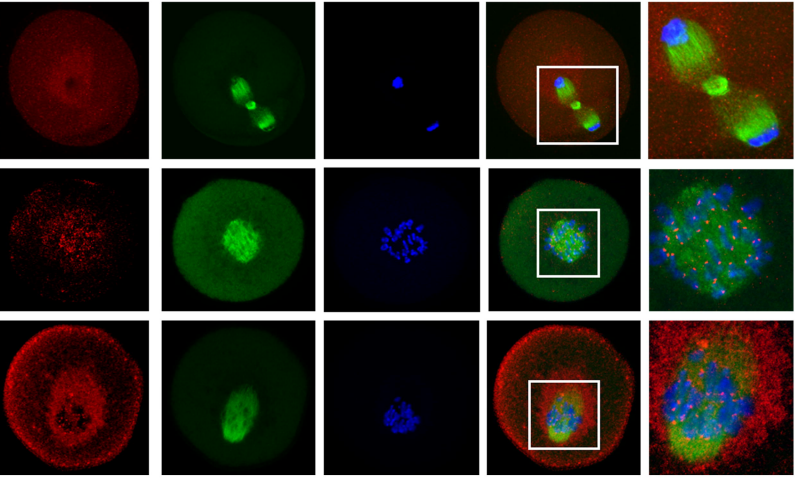
CDC6 regulates both G2/M transition and metaphase-to-anaphase transition during the first meiosis of mouse oocytes
Zi-Yun Yi, Tie-Gang Meng, Xue-Shan Ma, Jian Li, Chun-Hui Zhang, Ying-Chun Ouyang, Heide Schatten, Jie Qiao, Qing-Yuan Sun, Wei-Ping Qian
An autonomous oscillator times and executes centriole biogenesis
Mustafa G. Aydogan, Thomas L. Steinacker, Mohammad Mofatteh, Lisa Gartenmann, Alan Wainman, Saroj Saurya, Siu S. Wong, Felix Y. Zhou, Michael A. Boemo, Jordan W. Raff
Phase separation of zonula occludens proteins drives formation of tight junctions
Oliver Beutel, Riccardo Maraspini, Karina Pombo-Garcia, Cécilie Martin-Lemaitre, Alf Honigmann
Spatial integration of mechanical forces by alpha-actinin establishes actin network symmetry
Fabrice Senger, Amandine Pitaval, Hajer Ennomani, Laetitia Kurzawa, Laurent Blanchoin, Manuel Théry
Sumoylation regulates central spindle protein dynamics during chromosome segregation in oocytes
Federico Pelisch, Laura Bel Borja, Ellis G. Jaffray, Ronald T. Hay
Chromosomes function as a barrier to mitotic spindle bipolarity in polyploid cells
Alix Goupil, Maddalena Nano, Gaëlle Letort, Delphine Gogendeau, Carole Pennetier, Renata Basto
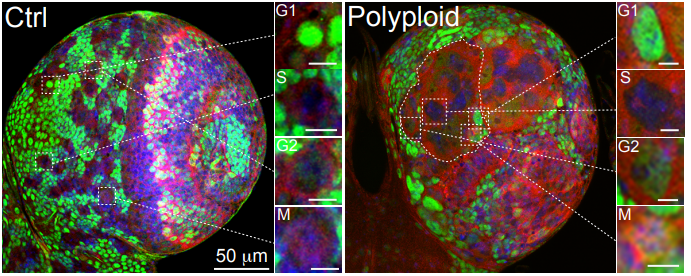
Cell Cycle Asynchrony Generates DNA Damage at Mitotic Entry in Polyploid Cells
Maddalena Nano, Anthony Simon, Carole Pennetier, Vincent Fraisier, Veronique Marthiens, Renata Basto
Cohesin and microtubule dependent mechanisms regulate sister centromere fusion during meiosis I
Lin-Ing Wang, Arunika Das, Kim S. McKim
Efa6 protects axons and regulates their growth and branching through eliminating off-track microtubules at the cortex
Yue Qu, Ines Hahn, Meredith Lees, Jill Parkin, André Voelzmann, Karel Dorey, Alex Rathbone, Claire Friel, Viki Allan, Pilar Okenve-Ramos, Natalia Sanchez-Soriano, Andreas Prokop
The model of local axon homeostasis – explaining the role and regulation of microtubule bundles in axon maintenance and pathology
Ines Hahn, André Voelzmann, Yu-Ting Liew, Beatriz Costa-Gomes, Andreas Prokop
A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance
Nicholas P Boyer, Laura E McCormick, Fabio L Urbina, Stephanie Gupton
Rho-dependent control of the Citron kinase, Sticky, drives midbody ring maturation.
Nour El-amine, Sabrya C Carim, Denise Wernike, Gilles R,X Hickson
Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell cycle arrest
Sangkyun Cho, Manasvita Vashisth, Amal Abbas, Stephanie Majkut, Kenneth Vogel, Yuntao Xia, Irena L. Ivanovska, Jerome Irianto, Manorama Tewari, Kuangzheng Zhu, Elisia D. Tichy, Foteini Mourkioti, Hsin-Yao Tang, Roger A. Greenberg, Benjamin L. Prosser, Dennis E. Discher
Vimentin protects the structural integrity of the nucleus and suppresses nuclear damage caused by large deformations
Alison E Patteson, Amir Vahabikashi, Katarzyna Pogoda, Stephen A Adam, Anne Goldman, Robert Goldman, Paul Janmey
Optogenetic control of cell morphogenesis on protein micropatterns
Katja Zieske, R Dyche Mullins
Dysregulated wild-type cell division at the interface between host and oncogenic epithelium
Megan Moruzzi, Alexander Nestor-Bergmann, Keith Brennan, Sarah Woolner
OVOL2 induces mesenchymal-to-epithelial transition in fibroblasts and enhances reprogramming to epithelial lineages
Kazuhide Watanabe, Ye Liu, Shuhei Noguchi, Madeleine Murray, Jen-Chien Chang, Mami Kishima, Hajime Nishimura, Kosuke Hashimoto, Aki Minoda, Harukazu Suzuki
β-Catenin tumour-suppressor activity depends on its ability to promote Pro-N-Cadherin maturation
Antonio Herrera, Anghara Menendez, Blanca Torroba, Sebastian Pons
Modelling
Cell signalling stabilizes morphogenesis against noise
Pascal Hagolani, Roland Zimm, Miquel Marin-Riera, Isaac Salazar-Ciudad
Evaluation of BMP-mediated patterning in zebrafish embryos using a growing finite difference embryo model
Linlin Li, Xu Wang, Mary C. Mullins, David M. Umulis
Notch signaling and taxis mechanims regulate early stage angiogenesis: A mathematical and computational model
Rocío Vega, Manuel Carretero, Rui D.M. Travasso, Luis L. Bonilla
A Monte Carlo method for in silico modeling and visualization of Waddington’s epigenetic landscape with intermediate details
Xiaomeng Zhang, Ket Hing Chong, Jie Zheng
A probabilistic atlas for cell identification
Greg Bubnis, Steven Ban, Matthew D. DiFranco, Saul Kato
The multicellular incoherent feedforward loop motif generates spatial patterns
Marcos Rodríguez Regueira, Jesús Daza García, Alfonso Rodríguez-Patón Aradas
Active Fingering Instability in Tissue Spreading
Ricard Alert, Carles Blanch-Mercader, Jaume Casademunt
Modelling adhesion-independent cell migration
Gaspard Jankowiak, Diane Peurichard, Anne Reversat, Christian Schmeiser, Michael Sixt
Stochastic nonlinear model for somatic cell population dynamics during ovarian follicle activation
Frédérique Clément, Frédérique Robin, Romain Yvinec
Bridging the gap between single-cell migration and collective dynamics
Florian Thüroff, Andriy Goychuk, Matthias Reiter, Erwin Frey
Bioelectrical signaling via domain wall migration
Harold M McNamara, Rajath Salegame, Ziad Al Tanoury, Haitan Xu, Shahinoor Begum, Gloria Ortiz, Olivier Pourquie, Adam Ezra Cohen
A moving grid finite element method applied to a mechanobiochemical model for 3D cell migration
Laura Murphy, Anotida Madzvamuse
Tools & resources
The protein repertoire in early vertebrate embryogenesis
Leonid Peshkin, Alexander Lukyanov, Marian Kalocsay, Robert Michael Gage, DongZhuo Wang, Troy J. Pells, Kamran Karimi, Peter D. Vize, Martin Wühr, Marc W. Kirschner
In vivo study of gene expression with an enhanced dual-color fluorescent transcriptional timer
Li He, Norbert Perrimon, Richard Binari, Jiuhong Huang, Julia Falo
An improved auxin-inducible degron system preserves native protein levels and enables rapid and specific protein depletion
Kizhakke Mattada Sathyan, Brian D. McKenna, Warren Anderson, Fabiana M. Duarte, Leighton J. Core, Michael J. Guertin
Unintended inhibition of protein function using GFP nanobodies in human cells
Cansu Küey, Gabrielle Larocque, Nicholas I. Clarke, Stephen J. Royle
Pervasive head-to-tail insertions of DNA templates mask desired CRISPR/Cas9-mediated genome editing events
Boris V. Skryabin, Leonid Gubar, Birte Seeger, Helena Kaiser, Anja Stegemann, Johannes Roth, Sven G. Meuth, Hermann Pavenstädt, Joanna Sherwood, Thomas Pap, Roland Wedlich-Söldner, Cord Sunderkötter, Yuri B. Schwartz, Juergen Brosius, Timofey S. Rozhdestvensky
CRISPR/Cas9-based mutagenesis frequently provokes on-target mRNA misregulation
Rubina Tuladhar, Yunku Yeu, John Tyler Piazza, Zhen Tan, Jean Rene Clemenceau, Xiaofeng Wu, Quinn Barrett, Jeremiah Herbert, David H. Mathews, James Kim, Tae Hyun Hwang, Lawrence Lum
Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing
Sundaram Acharya, Arpit Mishra, Deepanjan Paul, Asgar Hussain Ansari, Mohammad Azhar, Namrata Sharma, Meghali Aich, Dipanjali Sinha, Saumya Sharma, Shivani Jain, Arjun Ray, Souvik Maiti, Debojyoti Chakraborty
Analysis of single nucleotide variants in CRISPR-Cas9 edited zebrafish embryos shows no evidence of off-target inflation
Marie R Mooney, Erica E Davis, Nicholas Katsanis
An open-source and low-cost feeding system for zebrafish facilities
Astou Tangara, Gerard Paresys, Firas Bouallague, Yvon Cabirou, Jozsua Fodor, Victor Llobet, German Sumbre
Towards a fully automated surveillance of well-being status in laboratory mice using deep learning
Niek Andresen, Manuel Wöllhaf, Katharina Hohlbaum, Lars Lewejohann, Olaf Hellwich, Christa Thöne-Reineke, Vitaly Belik
A robotic platform for fluidically-linked human body-on-chips experimentation
Richard Novak, Miles Ingram, Susan Clauson, Debarun Das, Aaron Delahanty, Anna Herland, Ben M. Maoz, Sauveur S. F. Jeanty, Mahadevabharath R. Somayaji, Morgan Burt, Elizabeth Calamari, Angeliki Chalkiadaki, Alexander Cho, Youngjae Choe, David Benson Chou, Michael Cronce, Stephanie Dauth, Toni Divic, Jose Fernandez-Alcon, Thomas Ferrante, John Ferrier, Edward A. FitzGerald, Rachel Fleming, Sasan Jalili-Firoozinezhad, Thomas Grevesse, Josue A. Goss, Tiama Hamkins-Indik, Olivier Henry, Chris Hinojosa, Tessa Huffstater, Kyung-Jin Jang, Ville Kujala, Lian Leng, Robert Mannix, Yuka Milton, Janna Nawroth, Bret A. Nestor, Carlos F. Ng, Blakely O’Connor, Tae-Eun Park, Henry Sanchez, Josiah Sliz, Alexandra Sontheimer-Phelps, Ben Swenor, Guy Thompson II, George J. Touloumes, Zachary Tranchemontagne, Norman Wen, Moran Yadid, Anthony Bahinski, Geraldine A. Hamilton, Daniel Levner, Oren Levy, Andrzej Przekwas, Rachelle Prantil-Baun, Kevin K. Parker, Donald E. Ingber
CoBATCH for high-throughput single-cell epigenomic profiling
Qianhao Wang, Haiqing Xioong, Shanshan Ai, Xianhong Yu, Yaxi Liu, Jiejie Zhang, Aibin He
CellTag Indexing: genetic barcode-based sample multiplexing for single-cell genomics
Chuner Guo, Wenjun Kong, Kenji Kamimoto, Guillermo C. Rivera-Gonzalez, Xue Yang, Yuhei Kirita, Samantha A Morris
High-density spatial transcriptomics arrays for in situ tissue profiling
Sanja Vickovic, Goekcen Eraslan, Fredrik Salmen, Johanna Klughammer, Linnea Stenbeck, Tarmo Aijo, Richard Bonneau, Jose Fernandez Navarro, Ludvig Bergenstraahle, Joshua Gould, Mostafa Ronaghi, Jonas Frisen, Joakim Lundeberg, Aviv Regev, Patrik L Staahl
Mapping Histone Modifications in Low Cell Number and Single Cells Using Antibody-guided Chromatin Tagmentation (ACT-seq)
Benjamin Carter, Wai Lim Ku, Qingsong Tang, Jee Youn Kang, Keji Zhao
Estimation of cell lineage trees by maximum-likelihood phylogenetics
Jean Feng, William S DeWitt III, Aaron McKenna, Noah Simon, Amy D Willis, Frederick A Matsen IV
Characterizing the epigenetic landscape of cellular populations from bulk and single-cell ATAC-seq information
Mariano I. Gabitto, Anders Rasmussen, Orly Wapinski, Kathryn Allaway, Nicholas Carriero, Gordon J. Fishell, Richard Bonneau
A streamlined protocol and analysis pipeline for CUT&RUN chromatin profiling
Michael P. Meers, Terri Bryson, Steven Henikoff
Virtual ChIP-seq: predicting transcription factor binding by learning from the transcriptome
Mehran Karimzadeh, Michael M. Hoffman
Cell BLAST: Searching large-scale scRNA-seq database via unbiased cell embedding
Zhi-Jie Cao, Lin Wei, Shen Lu, De-Chang Yang, Ge Gao
Computational comparison of developmental cell lineage trees by alignments
Meng Yuan, Xujiang Yang, Jinghua Lin, Xiaolong Cao, Feng Chen, Xiaoyu Zhang, Zizhang Li, Guifeng Zheng, Xueqin Wang, Xiaoshu Chen, Jian-Rong Yang
Rosa26 docking sites for investigating genetic circuit silencing in stem cells
Michael Fitzgerald, Chelsea Gibbs, Tara Deans
Beyond early development: observing zebrafish over 6 weeks with hybrid optical and optoacoustic imaging
Paul Vetschera, Benno Koberstein-Schwarz, Tobias Schmitt-Manderbach, Christian Dietrich, Wibke Hellmich, Andrei Chekkoury, Panagiotis Symvoulidis, Josefine Reber, Gil Westmeyer, Hernán López-Schier, Murad Omar, Vasilis Ntziachristos
Linalool acts as a fast and reversible anesthetic in Hydra
Tapan Goel, Rui Wang, Sara Martin, Elizabeth Lanphear, Eva-Maria S. Collins

The mesoSPIM initiative: open-source light-sheet mesoscopes for imaging in cleared tissue
Fabian F. Voigt, Daniel Kirschenbaum, Evgenia Platonova, Stéphane Pagès, Robert A. A. Campbell, Rahel Kästli, Martina Schaettin, Ladan Egolf, Alexander van der Bourg, Philipp Bethge, Karen Haenraets, Noémie Frézel, Thomas Topilko, Paola Perin, Daniel Hillier, Sven Hildebrand, Anna Schueth, Alard Roebroeck, Botond Roska, Esther Stoeckli, Roberto Pizzala, Nicolas Renier, Hanns Ulrich Zeilhofer, Theofanis Karayannis, Urs Ziegler, Laura Batti, Anthony Holtmaat, Christian Lüscher, Adriano Aguzzi, Fritjof Helmchen
Scalable image processing techniques for quantitative analysis of volumetric biological images from light-sheet microscopy
Justin Swaney, Lee Kamentsky, Nicholas B Evans, Katherine Xie, Young-Gyun Park, Gabrielle Drummond, Dae Hee Yun, Kwanghun Chung
Digital removal of autofluorescence from microscopy images
Heeva Baharlou, Nicolas P Canete, Kirstie M Bertram, Kerrie J Sandgren, Anthony L Cunningham, Andrew N Harman, Ellis Patrick
Direct visualization of single nuclear pore complex proteins using genetically-encoded probes for DNA-PAINT
Thomas Schlichthaerle, Maximilian T. Strauss, Florian Schueder, Alexander Auer, Bianca Nijmeijer, Moritz Kueblbeck, Vilma Jimenez Sabinina, Jervis V. Thevathasan, Jonas Ries, Jan Ellenberg, Ralf Jungmann
Nuclear pores as versatile reference standards for quantitative superresolution microscopy
Jervis Vermal Thevathasan, Maurice Kahnwald, Konstanty Cieśliński, Philipp Hoess, Sudheer Kumar Peneti, Manuel Reitberger, Daniel Heid, Krishna Chaitanya Kasuba, Sarah Janice Hoerner, Yiming Li, Yu-Le Wu, Markus Mund, Ulf Matti, Pedro Matos Pereira, Ricardo Henriques, Bianca Nijmeijer, Moritz Kueblbeck, Vilma Jimenez Sabinina, Jan Ellenberg, Jonas Ries
PlotsOfDifferences – a web app for the quantitative comparison of unpaired data
Joachim Goedhart
colocr: An R package for conducting co-localization analysis on fluorescence microscopy images
Mahmoud Ahmed, Trang Huyen Lai, Deok Ryong Kim
ColourQuant: a high-throughput technique to extract and quantify colour phenotypes from plant images
Mao Li, Margaret H. Frank, Zoë Migicovsky
Facile assembly of an affordable miniature multicolor fluorescence microscope made of 3D-printed parts enables detection of single cells
Samuel B Tristan-Landin, Alan M Gonzalez-Suarez, Rocio J Jimenez-Valdes, Jose Luis Garcia-Cordero
Volumetric two-photon imaging in live cells and embryos via axially gradient excitation
Yufeng Gao, Xianyuan Xia, Jia Yu, Tingai Chen, Zhili Xu, Long Xiao, Liang Wang, Fei Yan, Zhuo Du, Jun Chu, Hairong Zheng, Hui Li, Wei Zheng
The Fruit Fly Brain Observatory: From Structure to Function
Nikul H. Ukani, Chung-Heng Yeh, Adam Tomkins, Yiyin Zhou, Dorian Florescu, Carlos Luna Ortiz, Yu-Chi Huang, Cheng-Te Wang, Mehmet K. Turkcan, Tingkai Liu, Paul Richmond, Chung-Chuan Lo, Daniel Coca, Ann-Shyn Chiang, Aurel A. Lazar
Dual-matrix 3D culture system as a biomimetic model of epithelial tissues
Diana Bogorodskaya, Joshua S. McLane, Lee A. Ligon
Research practice & education
The life of P.I. – Transitions to Independence in Academia
Sophie E Acton, Andrew Bell, Christopher P Toseland, Alison Twelvetrees
Breaking barriers: The effect of protected characteristics and their intersectionality on career transition in academics
Klara M Wanelik, Joanne S Griffin, Megan Head, Fiona C Ingleby, Zenobia Lewis
Comparing quality of reporting between preprints and peer-reviewed articles in the biomedical literature
Clarissa F. D. Carneiro, Victor G. S. Queiroz, Thiago C. Moulin, Carlos A. M. Carvalho, Clarissa B. Haas, Danielle Rayêe, David E. Henshall, Evandro A. De-Souza, Felippe Espinelli, Flávia Z. Boos, Gerson D. Guercio, Igor R. Costa, Karina L. Hajdu, Martin Modrák, Pedro B. Tan, Steven J. Burgess, Sylvia F. S. Guerra, Vanessa T. Bortoluzzi, Olavo B. Amaral
#Pay4Reviews: Academic publishers should pay scientists for peer-review
Rodolfo Jaffé
Ten myths around open scholarly publishing
Jonathan P Tennant, Harry Crane, Tom Crick, Jacinto Davila, Asura Enkhbayar, Johanna Havemann, Bianca Kramer, Ryan Martin, Paola Masuzzo, Andy Nobes, Curt Rice, Bárbara S Rivera-López, Tony Ross-Hellauer, Susanne Sattler, Paul Thacker, Marc Vanholsbeeck
Grant Reviewer Perceptions of Panel Discussion in Face-to-Face and Virtual Formats: Lessons from Team Science?
Stephen A Gallo, Karen Schmaling, Lisa A Thompson, Scott Glisson
Knowledge and attitudes among life scientists towards reproducibility within journal articles
Evanthia Kaimaklioti Samota, Robert P. Davey
Bob Patton
EvolvingSTEM: A microbial evolution-in-action curriculum that enhances learning of evolutionary biology and biotechnology
Vaughn S. Cooper, Taylor M. Warren, Abigail M. Matela, Michael Handwork, Shani Scarponi
Integrating case studies into graduate teaching assistant training to improve instruction in biology laboratory courses
Jelena Kraft, elise Walck-Shannon, Colleen Reilly, Ann Stapleton
Potential bias in peer review of grant applications at the Swiss National Science Foundation
Anna Severin, João Martins, François Delavy, Anne Jorstad, Matthias Egger
Why not…
Marc HE de Lussanet


 (No Ratings Yet)
(No Ratings Yet)