This month in preLights – September
Posted by preLights CoB, on 12 October 2018
Welcome to our monthly summary of developmental biology (and related) preLights.

It’s been almost eight months since the launch of preLights, and we are very excited about the hugely positive response from the community so far and preLights’ rapid growth – we recently reached two hundred preprint highlights! To further grow this service, we will be announcing a call for recruiting new team members next month, so watch this space if you would like to join us as a preLighter.
The preLights posts from September turned out to be very diverse: from worms and insects to plants and fish, and also plenty of exciting novel techniques. We hope you enjoy this selection!
From Development to Disease
The early C. elegans embryo is a powerful model to tackle questions like how polarity initially emerges in a developing organism. Giuliana Clemente covered two studies that revealed a new role for Aurora A in symmetry breaking and polarity establishment in the worm.
Sarah Bowling discussed why cells die when they are not in the “right place” and highlighted a preprint showing that disrupted EGFR signaling, rather than a fitness recognition event through cell competition, is responsible. The preprint authors gave further insight into the study (check out the author’s response section) and pointed out that the simplest explanation to their question proved to be the correct one.
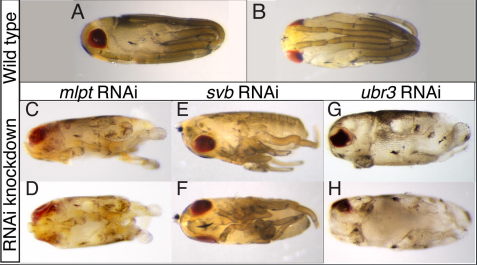
Erik Clark’s preLight showcased a conserved role for a posttranslational module (involving the micropeptide millepattes) in segmentation across many insect species, but a lack of segmentation phenotypes in Drosophila mutants. While they may not be good models for insect segmentation, Drosophila has provided numerous mechanistic insights into cell and tissue dynamics through the study of its developing egg chamber. Sundar Naganathan highlighted a preprint showing that in this model, 3D tissue elongation occurs through a mechanochemical signaling pathway that translates the external stiffness gradient into altered cadherin trafficking. Amanda Haage’s preLight also featured some fine mechanobiology in cancer cells, showing that actomyosin contractility regulates the activity of PKA, which is important for mechanically guided cell migration (durotaxis).
From the plant development field, Martin Balcerowicz wrote about a study showing that two new proteins (Sol1 and Sol2) regulate fate transitions and cell divisions during Arabidopsis stomatal development.
Moving to zebrafish, Alberto Rosello-Diez featured a novel mechanism that initiates regeneration upon injury: in response to ROS signaling, Hegdehog-expressing cells are repositioned to create a new signaling center. Andreas van Impel reviewed zebrafish vasculature development and how the 50:50 balance of intersegmental arteries and veins is regulated by a flow-dependent mechanism. Excessive blood vessel formation, on the other hand, is a hallmark of cancer; Coert Margadant highlighted a promising strategy to inhibit blood vessel formation by blocking recycling of phosphoinositide lipids.
Several preprints harnessed the power of single-cell transcriptomics to gain insight into neural development and neurodegenerative disease. Zheng-Shan Chong covered a study exploring microglial development through scRNA-seq, which discovered a subset of postnatal neural cells resembling degenerative disease-associated microglia (DAM). Yen-Chung Chen preLighted two preprints looking at why not all neuron types are equally vulnerable in neurodegenerative diseases. Applying new transcriptomic techniques, LCM-seq and Axon-seq, potential protective mechanisms utilized by resistant somatic neurons were found. Finally, Carmen Adriaens’ preLight also dealt with high-throughput sequencing (promoter capture Hi-C), and explained the links between SNP-s associated with type 2 diabetes and chromatin architecture of human pancreatic islet cells.
Tools & Technologies
It’s difficult to imagine where the developmental biology field would be today without in situ hybridization techniques that detect mRNA localization. This month, Yen-Chung Chen highlighted work that developed a new fluorescent in situ hybridization method termed SABER, which is more sensitive, cheaper and quicker compared to previous FISH methods. To look at fluctuations in mRNA levels and whether these persist over multiple cell divisions, the preprint covered by Leighton Daigh combined Luria and Delbrück’s fluctuation analysis with population based mRNA sequencing (this new method was termed MemorySeq).
The number of preprints dealing with CRISPR technology is rapidly growing, but there is still a lot to be understood regarding the targeting and editing accuracy and efficacy in this system. Here, Rob Hynds preLighted a study that uncovered some general principles of CRISPR gene editing by analyzing a large set of indel profiles.
Jacky Goetz wrote about a new method that is able to quantify mechanical stress in vivo by injecting polyacrylamide beads into living embryos and analyzing their deformations. Finally, one of the favorites from this month was Samantha Seah’s preLight that discussed a new way of visualizing data using raincloud plots.


 (1 votes)
(1 votes)