Using RNA sequencing to understand real-world embryos
Posted by Elin Sørhus, on 22 March 2017
Comment on “Novel adverse outcome pathways revealed by chemical genetics in a developing marine fish” eLife 6 (2017)
Elin Sørhus, John P Incardona, Tomasz Furmanek, Giles W Goetz, Nathaniel L Scholz, Sonnich Meier, Rolf B Edvardsen, Sissel Jentoft
Institute of Marine Research, Norway
Northwest Fisheries Science Center, USA
University of Oslo, Norway
Crude oil contains many chemicals which are toxic to both humans and aquatic animals. Fish embryos, in particular, are sitting ducks when it comes to oil pollution. The vast majority are deposited naked, unprotected by their mother, drifting around or stuck in a place on some plants or rocks. What happens if they drift through an oil spill or are deposited next to an oiled beach?
Oil spills are happening all the time. Well known large spills that got a lot of publicity are the Exxon Valdez oil spill in Prince William Sound in Alaska in 1989, which affected local animal populations for decades, and the Deepwater Horizon wellhead blowout disaster in the Gulf of Mexico in 2010, where we probably still don’t see the full consequences. However, we also find examples of recent spills that get very little worldwide attention. For example, there was a major oil spill near Chennai in India in February 2017 with a release of nearly 20 million litres of heavy bunker fuel oil. This latter spill was not mentioned in any European newspapers, suggesting there are more black oily numbers out there. Understandably, oil spills create a great concern for long-term effect on the environment, especially for fish spawning areas where eggs are released, and we search for tools to predict and measure these consequences.
The Deepwater Horizon disaster has really intensified the amount of studies investigating developmental abnormalities in fish induced by oil exposure. These studies identified the developing heart as the major target of crude oil. Exposure results in both functional and developmental abnormalities by directly interfering with ion currents in heart muscle cells (cardiomyocytes). This means that in addition to immediate effects on function during exposure, an altered heart shape resulting from an early life exposure also can generate a reduced cardiac output later in life. Reduced cardiac performance in turn will diminish swimming ability, potentially decreasing feed intake, predator avoidance and survival. For example, a reduced cardiac output in juvenile fish exposed to oil as embryos may have contributed to the collapse of the herring stock in Prince William Sound in Alaska a few years after the Exxon Valdez oil spill.

I work for a government institute that manages Norway’s marine resources, most importantly the fish stocks that are a major part of our economy. As an applied science agency, we need to understand how Norway’s other big economic activity, offshore oil production, potentially affects the fitness of our fish stocks. Crude oil’s effect on the heart and potential impacts on juvenile survival were therefore the primary focus when we performed our first experiment in 2013. At the time, I was a newly hired PhD student thirsting for challenges. I had just left my position as a lab technician with 10 years experience in the vast field of molecular biology. As a student in a Genomics program, I was recruited to perform RNA sequencing studies in oil-exposed marine fish with our institute’s Ecotoxicology group. The goal was to relate changes in the whole-animal embryonic and larval transcriptomes to developmental defects. But I was the rookie and totally a blank slate when it came to crude oil toxicity and developmental abnormalities. Fortunately, I was born into this world with an exceptional overload of energy which is easily transferred to hard work, long hours and optimism despite some downfalls. Also, past experience with experiments that did not turn out as expected taught me efficient trouble shooting, and when to change course and to easily create solutions to unexpected problems.
We started out working on embryos of Atlantic haddock (Melanogrammus aeglefinus) because it is an important fisheries resource in Norway and the rest of the North Atlantic. The main spawning grounds for haddock in Norway are targeted for expanded offshore oil production. In the first year our plan was to do two experiments, exposing early embryos and newly hatched larvae to dispersed crude oil. However, dealing with a non-model marine fish broodstock, getting embryos at the right time wasn’t so simple. Embryos were produced before we were really ready, but we managed to pull off an experiment exposing embryos after completion of major organogenesis. Even though this wasn’t the ideal experiment, we made a very striking and disturbing observation. Oil droplets stuck to the apparently relatively sticky chorion of haddock eggs. Presumably this increased and prolonged their exposure. Thus, despite not being exposed at their most vulnerable stages of development, these embryos still showed surprisingly high deformity rates by the time they hatched. This included large amount of edema formation in the heart sac, which is indicative of heart failure. But we weren’t able to assess these defects in sufficient detail.
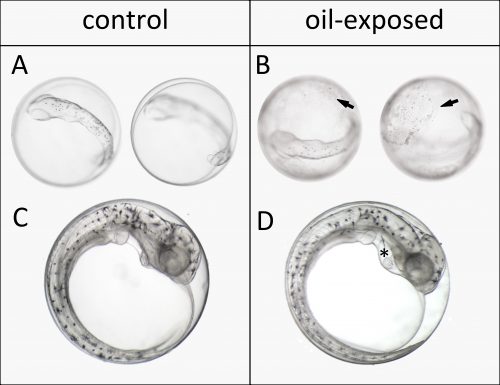
To assess these effects more thoroughly, the year after, we invited John Incardona and his group to participate in the experiments. As a developmental biologist with over a decade of oil exposure experience, John taught us how to perfectly position and video image these delicate creatures. He showed us excitedly almost invisible development markers in our tiny haddock larvae, like the urinary tract and lateral line neuromasts. For me, imaging and sampling day in and day out, I dreamt about beating fish hearts and doing the swoop through methylcellulose to position the larvae in my sleep. Still, in the end I regret I didn’t take that extra shot. Nevertheless, I somewhat unknowingly created a sufficiently large and detailed picture and video library during development, which proved to be crucial for interpreting the RNA sequencing data.

An applied science issue for natural resource agencies like ours is that there are too many species in the wild to develop species-specific tools on the level of model species like zebrafish. Working with non-model species like Atlantic haddock, for example, in situ hybridization probes and protocols are not readily available. Designing suitable species-specific assays and probes is time consuming and often very difficult because of lack of optimized methods and genome sequence knowledge. However, most of the fish species we care about, including haddock, have translucent embryos. The embryos therefore often can tell us directly what is happening with their development, if it’s normal or abnormal. We exploited this benefit when we used RNA sequencing to study normal development, linking changes in developmental processes and appearance of structures to temporal mRNA levels in a previous paper (Sørhus et al., 2016), showing that for example cardiac determination and differentiation genes preceded expression of genes related to cardiac function. However, would we be able to “see” abnormal structures represented in a temporal snapshot of mRNA levels? We know that altered structures typically have different distributions of cell types, theoretically this should be reflected in different levels of gene expression corresponding to cell type-specific markers. One example can be found in John´s early work (Incardona et al., 1998). He showed that the neural tube in embryos are mis-patterned after exposure to hedgehog-blocking teratogens. One pool of cell-types was replaced by another, and was measured by counting cells using antibodies for cell-type specific protein markers. If we can count cells this way, we should also be able to quantify them by mRNA sequencing. One important assumption however, is that mRNA transcript levels reflect protein levels. This is generally true in developing embryos, where genes are being turned on and off because gene function needs to be dynamic. Over-expression studies support this: generally if a cRNA is injected into an embryo, it makes a functional protein.
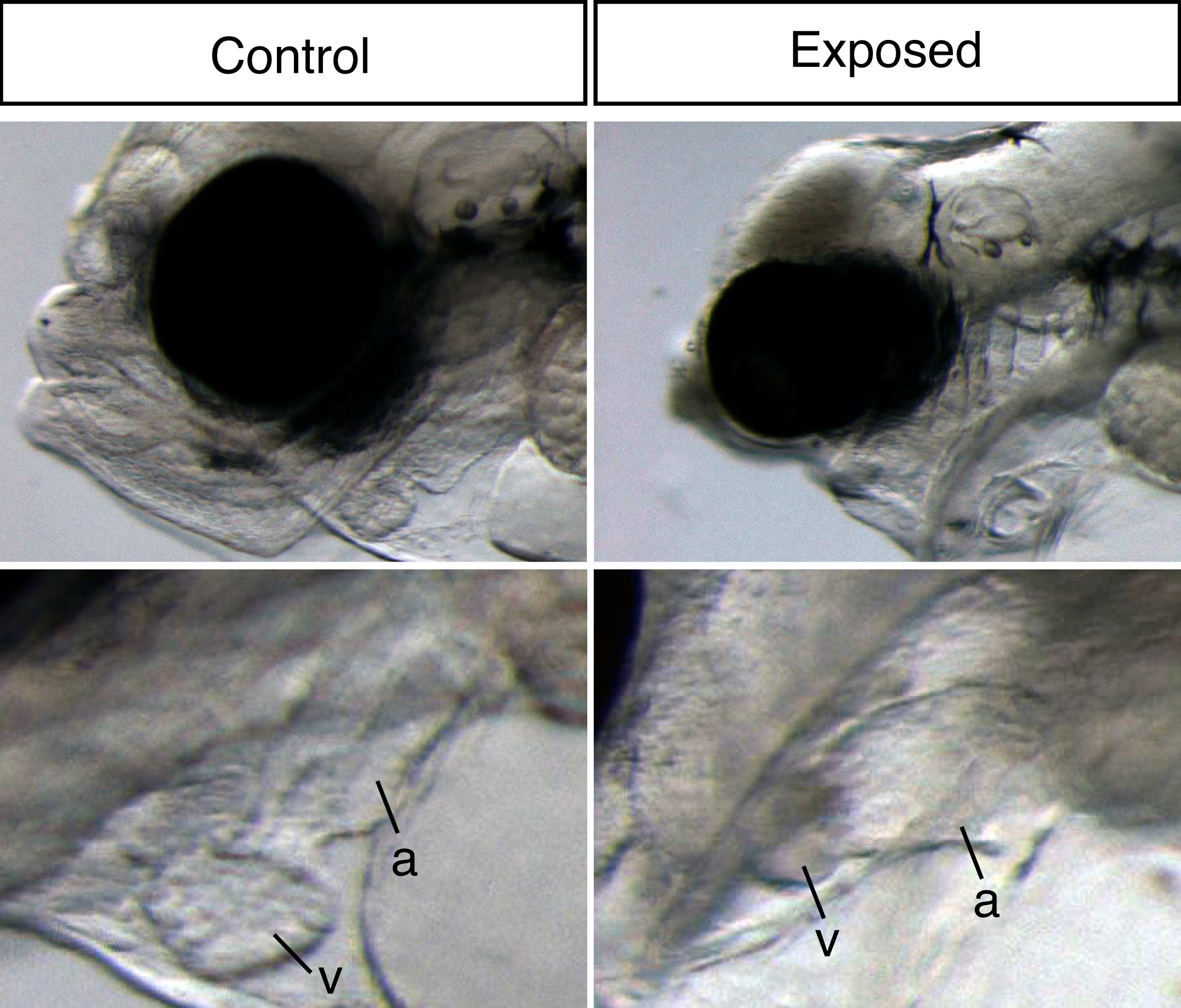
Our large picture and video library monitoring abnormal development almost continuously through the exposure experiment gave us a unique possibility to link gene expression changes to visible phenotypes. Thus, our main aim was to link gene expression to oil induced phenotypes and ultimately identify initiating events potentially leading to these phenotypes. The ultimate goal is to create a toolbox of molecular indicators that can help predict the developmental consequences and thus survival after either accidental real life oil spills or chronic low-level oil pollution. With both the RNA sequencing and image data wrapped inside our computers, the newbie (myself) and the expert (John) started a year long interpretation journey.
When dealing with 20 000 genes and 11 developmental stages, the use of bioinformatics tools is, of course, inevitable. This included the use of tools developed in-house as well as several commercially available pathway analysis databases. These tools are highly based on the biomedical literature focusing on mammalian species, and thus, do not necessarily represent the same processes in a marine fish species. So while they are quite powerful at identifying altered pathways, the challenge is figuring out which of those altered pathways are relevant to phenotypes. To avoid potential pitfalls of these bioinformatics pathway tools and not use them simply as a meat grinder, we therefore manually inspected individual genes underlying affected pathways. For example, in our oil exposed haddock, “salivary gland” was a pathway that came up as affected, which is quite peculiar considering fish do not produce spit. However, when inspecting the underlying genes, we saw that they were clearly involved in ion and water balance, which fit our oil exposed phenotype perfectly. For the same reason, we also performed our own “pathways analysis” the poor man’s way. This meant (simply!) diving into literature of developmental biology and studying known mutant phenotypes resembling our oil exposed fish larvae, making our own massive lists of genes involved in, for example, craniofacial and cardiac development, tissues that visually were highly affected in oil exposed haddock. For me, this was like venturing into the deep end of the pool.
A major problem we grappled with was how to determine whether individual genes of interest were the correct tissue-specific isoform. In other words, how could we have some degree of confidence that tissue-specific genes could be seen in the pool of mRNA extracted from whole embryos. Although we experimentally lacked information about localization of expression, underway we realized how to exploit the RNA sequencing data for this information. What bioinformatics pathway analyses don’t do is account for expression patterns, they generally rely on genetic interactions identified in the literature. We therefore turned to expression pattern databases in zebrafish (like Zfin.org). Based on known mRNA localization from other species like zebrafish, we could correlate the number of transcripts determined by the RNA sequencing with tissue-specific patterns at any particular time point. For example, genes shown to be expressed only in the developing heart in zebrafish all had transcript numbers in a limited range. Thus, if a gene of interest had transcript levels well above that range, we knew it had to have a broader expression pattern. Similarly, pathway knowledge and mutant studies in the literature showed us what to expect for the down-stream consequences of an abnormal up or down-gene regulation. When manually curating gene pathways of craniofacial and cardiac development, this information was always in our minds.
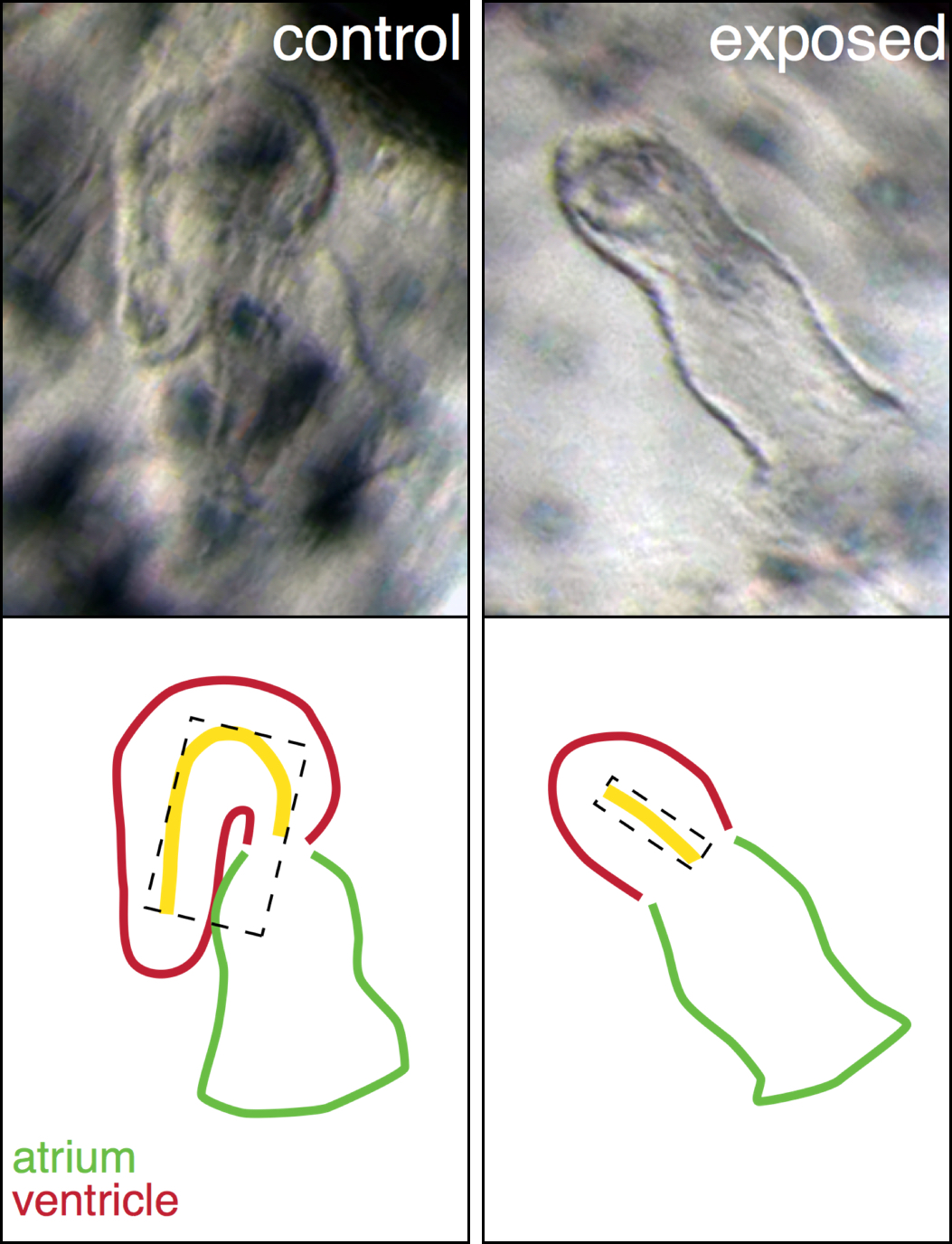
A perfect example is our explanation for the abnormal heart shape in oil exposed haddock. Most of the hearts in the highest exposure group were un-looped, with the atrial and ventricular chambers in a linear rather than side-by-side arrangement, and the ventricle was extremely underdeveloped. From a large list of genes involved in cardiogenesis, one gene, bmp10, stood out and was abnormally regulated at one of the earliest time points. This gene was therefore a strong candidate for one of the first transcriptional events leading to the abnormal heart development. Bmp10 is a strong morphogen that is normally involved in the trabeculation process, the outgrowth of fingerlike structures in the ventricle. However, trabeculation is a process that is occurs much later in heart development, around hatching stage in fish. Remarkably, we detected bmp10 up-regulation when the heart was just at the “cone” stage, before the chambers are really separated and cardiomyocytes are even contracting. At this stage, another Bmp gene is normally expressed, bmp4, and it is needed to lateralize the cone and drive the asymmetric looping process to bring the chambers side-by-side. Functional studies in zebrafish demonstrated that, as a potent morphogen, the activity of bmp4 must be tightly regulated, otherwise either too much or too little leads to an un-looped heart. Therefore, inappropriately turning on the bmp10 signal too soon most likely overrides the normal bmp4 signal, leading to an abnormal, un-looped heart. Downstream targets of Bmp10 were also up-regulated such as nkx2.5, a key transcription factor in cardiac development. An inappropriate overexpression of nkx2.5 yields a reduced ventricle in zebrafish, just like the one we see in our oil exposed haddock.
The bmp10-story is an excellent example of the hypothesis-based search for changes in gene expression linked to a visible phenotype. However, an advantage of unbiased RNA-sequencing dataset is the opportunity to conduct analysis where no main hypothesis is fully settled. The differentially expressed genes and pathways pointed to phenotypes of which we were peripherally aware, but didn´t pay strict attention to. This leading us to also focus on edema accumulation in the context of abnormal osmoregulation and linking disordered lipid metabolism and up-regulation of cholesterol synthesis to reduced yolk absorption.
Real life accidental oil spills are probably more common than we fear. The fact that an oil spill with 20 million litres of oil spread on a shore in India without being mentioned in the European media is a scary example. Therefore, it is more important now than ever to understand the underlying mechanisms of crude oil toxicity. In this study, we provide a toolbox of tissue specific biomarkers that together might be included in an important monitor apparatus in risk and damage assessment in the future. Which means that in the time ahead we can collect samples after accidental oil spills, like the one in India, Gulf of Mexico or Alaska, and predict the developmental effects and thus the consequences for the individual and eventually the population in the affected area. On the other hand, our results provide an improved general understanding about low-level, complex mixture toxicity. Even mammalian embryos and fetuses are likely impacted by the vast array of chemicals we dump into the environment in ways that we don’t currently understand. Thus, the data in this eLife paper is also generally important, even for people.
References
Incardona, J.P., Gaffield, W., Kapur, R.P., Roelink, H., 1998. The teratogenic Veratrum alkaloid cyclopamine inhibits Sonic hedgehog signal transduction. Development 125, 3553-3562.
Sørhus, E., Incardona, J.P., Furmanek, T., Jentoft, S., Meier, S., Edvardsen, R.B., 2016. Developmental transcriptomics in Atlantic haddock: Illuminating pattern formation and organogenesis in non-model vertebrates. Dev. Biol. 411, 301-313.


 (7 votes)
(7 votes)