BSDB Gurdon Summer Studentship Report (2)
Posted by BSDB, on 27 November 2014
In 2014, the BSDB has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. The 10 successful applicants spent 8 weeks in the research laboratories of their choices, and the feedback we received was outstanding. Please, read the student report kindly sent to us by George Choa.
Is there such thing as a housekeeping gene?
This summer I undertook an internship in the Research Department of Cell and Developmental Biology (CDB) at University College London (UCL). Joining the Organiser subgroup, my project involved determining the existence of a “housekeeping” gene (HKG), a gene that has both ubiquitous and uniform expression across different tissues types, no matter normal or diseased tissue, no matter the stage of embryo development, and no matter the model system used. HKGs act as reference genes in cases where magnitude of expression is required to be normalised, for example in RT-PCRs and northern blotting.
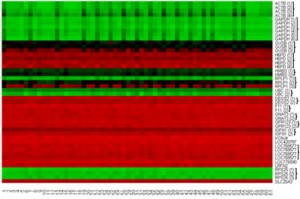
In order to confine the number of candidate genes for the project, specific criteria were applied to two sets of chick RNA data – quantitative expression values from microarray screens and RNASeq tissue assays, which were collected over time at both the Stern Lab at UCL and the Streit Lab at King’s College London (KCL). Simply done on Microsoft Excel, variance for each mRNA was calculated across samples of a variety of chick tissue types and stages, to which they were ranked. The least variant 5% (P = 0.05) and 1% (P = 0.01) mRNAs from microarray and RNASeq data, respectively, were selected. As this still left us with a total of more than 1500 mRNA HKG candidates, further filtration was applied, for example comparing the candidates to existing results in expression databases such as ZFin. This criteria resulted in a selection of 14 candidate HKGs, including genes that encode ribosomal proteins (RPS25) and channel proteins (KCNJ4). On top of this, an additional 9 probes that are considered conventional HKGs were also tested to confirm their reliability as HKGs; these conventional genes are commonly used as a baseline for a multitude of developmental studies, for example GAPDH and ACTB. All these probes were presented by box plots and heat maps using R, composed by me, taught by the Stern lab bioinformatician at the time; these were used in my end-of-project presentation.
 The more practical side of my project came next. My first week consisted of firstly learning to harvest chick embryos. This required a lot of patience and resolution, something I’ve come to appreciate to be two of the most important features of working as a researcher in science. One of the biggest challenges was the application of different techniques when harvesting embryos of different stages; some could be harvested quickly, most others took time and extreme delicacy. It was easy competing with myself from the previous day as each day I matured my technique, collecting dozens in one sitting, easily collecting over 150 different embryos of different Hamburger Hamilton (HH) stages over my first six weeks.
The more practical side of my project came next. My first week consisted of firstly learning to harvest chick embryos. This required a lot of patience and resolution, something I’ve come to appreciate to be two of the most important features of working as a researcher in science. One of the biggest challenges was the application of different techniques when harvesting embryos of different stages; some could be harvested quickly, most others took time and extreme delicacy. It was easy competing with myself from the previous day as each day I matured my technique, collecting dozens in one sitting, easily collecting over 150 different embryos of different Hamburger Hamilton (HH) stages over my first six weeks.
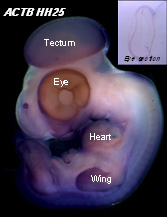
Following learning the tricks of the embryology trade, I dove into the more molecular side of my project – in situ hybridisation (ISH). Probes were synthesised and purified, and harvested chick embryos were processed, both by Stern lab protocols, ready to be stained. From this, the probes that appeared convincingly uniform in expression, as well as a variety of the conventionally used HKGs, were processed through wax sectioning to gain a better, more comprehensive look at the extent of the staining, before, and after, which they were documented.
A range of data (115 embryos) were collected by the end of my project, and with the several-odd wax section images, I found that none of the candidate genes matched the requirements of a HKG; even those that were sectioned due to convincing ubiquitous expression, which includes those conventional HKGs that hitherto are being used to normalise expression data.
Having collected all my data and completed my project to the best of my abilities, I presented my work in one of the many lab meetings I had the opportunity to attend. It was without doubt odd to sum my eight weeks spent researching into a 15 minute PowerPoint, but it was also wholly rewarding to see all I accomplished over my time at the Stern lab. Data collected from my project seem quite cogent in that there is probably no such thing as a HKG, which the lab hopes to publish in the near future.
Going into my final year of my Biomedical Sciences degree, I cannot thank everyone in the lab enough for the guidance, support and endless conversation that kept conducting my project so lively and enjoyable. I must however thank in particular my principal investigator Claire and Tutor Claudio for being so patient with me throughout the whole endeavour, and I hope one day I will return to the lab to take on my own PhD.
George Choa, BSc Biomedical Sciences, UCL



 (7 votes)
(7 votes)
Whoa! Very interesting! Congratulations, George, for such an interesting first step in research
Great to see that the Gurdon Summer Projects are already having a great impact by allowing several students to take on this opportunity of experiencing a hands-on summer in the lab! Keep it going, it’s great to see the initiative thrive!