An interview with Eric Wieschaus
Posted by the Node, on 18 May 2017
I had started to become a little worried when I didn’t see Eric on the opening day of the conference, but it turned out that his plane to Germany had been delayed by the snowstorms blanketing the Eastern seaboard of the US and he made it in the end. Between sessions later on, we found a slightly chilly empty computer lab to sit down in, and over the course of an hour talked about his history in research and views on science in general. My lasting impression was of an enthusiasm and passion for research that was by equal measure refreshing and infectious – it made me want to look down a microscope again!
This interview by Aidan Maartens first appeared in Development, Volume 144, Issue 10.
Eric Wieschaus is a HHMI Investigator, and the Squibb Professor in Molecular Biology at Princeton University and the Lewis-Sigler Institute for Integrative Genomics. In 1995, he was awarded the Nobel Prize jointly with Edward Lewis and Christiane Nüsslein-Volhard for discoveries about the genetic control of Drosophila embryogenesis. We caught up with Eric at the joint meeting of the German and Japanese Societies of Developmental Biologists held in Kiel in March 2017, and discussed his career, his thoughts on the field and the impact the Nobel award had on his life.

I understand that after your first experience with Drosophila – as an undergraduate in Harvey Bender’s lab at Notre Dame – you said never wanted to see a fly again! What led you to change your mind?
I suppose the formative undergraduate experience wasn’t washing dirty fly bottles and making fly food, which is what I had done in Harvey Bender’s lab, but an embryology course where we looked at frog and chick embryos, which were the embryos people worked with at the time. I knew then that I wanted to be an embryologist, to study development. But I think I just didn’t know that flies had embryos! Anyway, I ended up at Yale in New Haven, and for various reasons I was assigned to the lab of Donald Poulson. Poulson had, in the 1930s and 1940s, first as a graduate student and through to his professorship, described all of Drosophila embryogenesis. So I knew that I wanted to study embryos, and I had by chance landed in the laboratory of the one person in the world who really knew how Drosophila embryos develop. Poulson was a very kind person: he took me in to his lab when he specifically didn’t want to have a graduate student. It took him a little while to find a young assistant professor who was willing to take me on, and that turned out to be Walter Gehring.

And as a graduate student with Gehring, you were trying to map cell lineage in Drosophila with somatic clones and disc transplantation. What was the drive behind this work?
Walter had developed a tool to culture embryos, based on the strategies people had used to culture imaginal discs. He was able to convince me, a naïve young graduate student entering the lab, that the best experiment in the world would be to isolate a single cell from the blastoderm, grow it in these culture conditions and measure its developmental capacity, to establish whether it had already undergone some determination or programming. And that was my project – I spent my first three years grinding up embryos and trying to get single cells to grow. I have to say that none of that ever worked, but at some point I decided that I needed to have a control – once I had these wonderful cultures going, and these cells displaying their inherent potential, I wanted to be able to compare them to what a blastoderm cell actually did if left in situ. I decided I would produce clones at the blastoderm stage and follow those cells, but I never really looked at them because I still believed my experiments would work. But at the end of four and a half years, when I realised that I wasn’t going to have cultured cells as part of my thesis, I decided to go back and look at the controls. Luckily, it turned out that what cells normally do in development is almost as interesting as what you can get them to do when you artificially manipulate them.
When Walter’s lab moved to Basel, you were brought into contact with Christiane (Janni) Nüsslein-Volhard. How did that work out?
Janni and I both finished our thesis work at around the same time – she was in Tübingen, and having done molecular biology was an attractive person for Walter to bring to his lab. She actually came to Basel specifically not to do molecular biology, but instead to learn how to look at embryos, and as I was the only person in the lab who was working with embryos, we bonded over that in the four months before I left. And later, we both were fortunate to get positions in Heidelberg – these were real group leader positions, but we didn’t know that we had to bargain for space! So we ended up together, sharing a lab, and we already knew kind of what we wanted to do; it was a perfect set of circumstances.

And in Heidelberg, you began work on the screen that would eventually win you the Nobel Prize. Screens obviously require a lot of time and effort: how did you stay focussed and cope with the practical demands at the time?
Well it was a lot of work, but the payoffs were real. We spent a certain amount of time figuring out how to do the screen, and that was maybe more frustrating than actually doing it. But once we started the big screen, scaled up and lasting about two and a half months, we discovered something new every day, and so focus was not at all hard. We would work from nine in the morning to around midnight, pretty much every day of the week, but the payoff was already there – not in understanding as such, but in the realisation that we were seeing stuff that no one had ever seen before, and thinking thoughts that no one had ever thought before. I don’t even remember being frustrated or wanting to slow down at all – my memory of that time is just of the fascination and excitement of doing it.
We were seeing stuff that no one had ever seen before, and thinking thoughts that no one had ever thought before
Was there a key lesson the screen taught you about how development works?
Before we did the screen, we really didn’t know what we would get out of it. The two dangers were that every gene that you mutated would mess up development, and that the phenotypes would be complicated and heterogeneous and impossible to interpret. But what actually happened was that the number of zygotically active genes we found was small – about 120 to 130 – and the mutants had unique phenotypes that identified specific processes, which meant you could immediately group things. This small number contrasted with the characterisation of RNA heterogeneity carried out at the time, which estimated there were thousands of different mRNAs present in the embryo, but most of these turned out to come from the mother. So the conceptual lesson was that the embryo uses transcription of a small number of genes to drive specific events forward. If gene products can be supplied to all cells, they’re provided maternally, but if you want to turn the gene on at a specific place or at a specific time, zygotic transcription does that. That was the power of the screen, to pick up these regulators: the genes controlling developmental decisions. It seems quite obvious now, but did I predict that at the time? Absolutely not.
Following the screen, you moved your lab to Princeton in 1981. What did you hope to achieve in the early days in New Jersey?
Initially, when we returned to Princeton, I helped Trudi (Schüpbach) a bit when she was setting up her maternal effect screen to find the determinants deposited in the egg by the mother. But what I really wanted to understand was morphology, and this very quickly became a cell biological problem, of how cells move and change shape and so on. This obviously required a lot of observation, and I remember at the time the great discovery was that there was something called phalloidin (which labels F-actin) – it was fluorescent, and you could buy it from a supplier! You could get a fluorescence microscope and just look – it really allowed you to see embryogenesis differently. So this period was also a time where, as well as advances in molecular biology, there were great advances in cell biological reagents, which you could begin to apply to embryos to think about these processes from a more cell biological perspective. It all came together – the observation of the real things that cells do, and their manipulation by genetics (a tool that cell biologists did not have at the time).
And how did you approach the cell biological side of development, particularly since many of the factors involved would have been missed by the screen?
The screen picked switches, decision-making mechanisms: most of these are transcription factors and the cell signalling pathways that govern them. Once you go downstream of decisions, it becomes more complicated and more synthetic, and less easily approachable by a genetic screen that looks for survival, for instance. I think that genetic manipulations are still really powerful here, but you can’t take shortcuts like only looking for genes essential for viability. A number of biological processes go into a change in cell shape, for example, and if you remove one of them, the cell shape transition may not occur in the same way, but the animal may be robust enough that a proportion of embryos survive and you’ll miss it. So understanding morphology turns out to be a process where you use genetics, but need other tools too, and much of this concerns imaging. I think that a major transformation in biology has been the development of microscopes of different kinds, whether confocal or light sheet, or whatever. And digital cameras! To those of us who can remember, images used to be things you had on film and you would spend artistic moments in the darkroom trying to bring out what you wanted; now they are just a matrix of pixels. And I think that aspect – doing microscopy on embryos, and doing it in a quantitative way – meant you learned stuff that you didn’t know before. It’s just transformed the field.
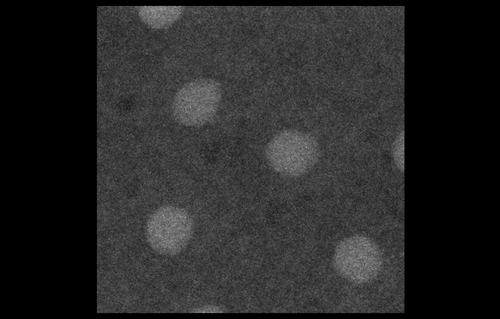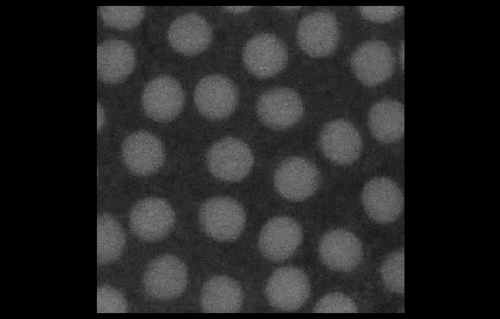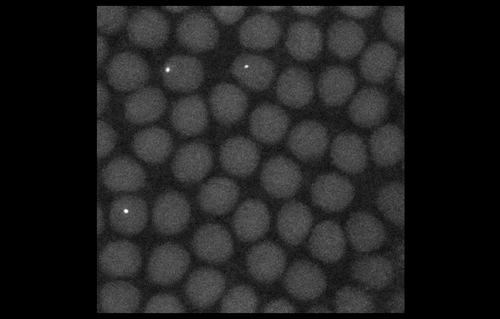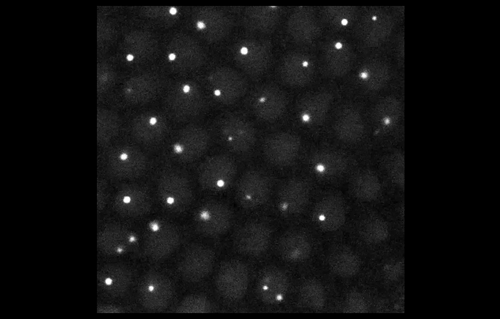
To look at embryos with a quantitative eye, your lab has in recent years incorporated physics and mathematical modelling. From your background, to what extent do you personally need to understand the physics and the maths?
Well I’m totally dependent on having collaborators who have the patience to explain things to me three or four times, and also the tolerance to accept when I decide that I’ve got it, even though there might be a deeper level of understanding available that they feel I haven’t reached yet. But it’s also an interesting question to ask what your thresholds are for understanding: I think you set them, and that sets the style of science that you do. The fact that different scientists have different thresholds is one of the reasons why it is so valuable to collaborate, and also why the social nature of science is so important to its productivity. You profit from talking to people who are interested in working with you, but frankly not willing to invest the same amount of energy into your topic as you do. Of course there’s always danger in not understanding everything, but we live in a dangerous world. Scientifically, if you are going to be on the edge of discovery, you just take the best path forward even if you don’t understand everything in a deeply defendable way.
As someone who has worked on Drosophila for 40 years, what do you think this animal still has to teach us about development?
Science is hard, and in biology we are only at the beginning of a truly deep understanding. To understand development, you work on embryos, and you choose a model organism because to do things well, you need the technologies that everybody else has developed; you need to parasitise a field.
Working with Drosophila today, you have two avenues. If you want to strike out into some totally unexplored area, for instance in something behavioural or evolutionary, flies allow you to do this. And then there are the old areas, things that we think of as established and successful problems, like patterning in the early embryo. What we have there is actually a wonderful cartoon understanding of the process, and this is so rare and valuable in biology because it can provide the basis for building a deeper understanding. So if you want to understand transcription, for example, I think you’d be crazy not to work in flies! In flies you have this organism where you can study a real process with biological meaning that has been selected for in evolution, and which is primed by our existing knowledge for you to start. So the things that we know prime us to go deeper, and for the things that we don’t know, we’re in a position where we can profit from the community and the tools that are available.
In biology we are only at the beginning of a truly deep understanding
It’s now 22 years since you won the Nobel Prize along with Christiane Nüsslein-Volhard and Edward Lewis. What did the award mean to you at the time, and does it still have an impact on your academic life?
Of course it was a wonderful thing to happen for me and my family. It wasn’t really anything I’d foreseen, and then I got ripped out of my normal life for a week in Stockholm. But I also felt like it was wonderful for the community as a whole: at least from my perspective, my colleagues were happy because they saw it as recognition of a set of accomplishments that the field had made.
The other thing that it gave me was a certain power to control my work life. I’ve never assumed major administrative roles at the university – it gave me the power to insist that I would continue to work four hours a day at the bench myself, and meant that people were happy to have me around even if it forced some others to assume extra responsibilities. I could set certain standards for my life: I wanted to be a productive scientist, not to just run a lab and tell other people what to do. I wanted to be the person who did interesting things.

You’ve described yourself as a very visually oriented person, both in terms of science and as someone who loved to draw and paint in your youth. Does this dictate the way you do your science?
Since the public is giving their own money to support science, there are limited resources, an obligation to produce and a certain competition among scientists. So as a young scientist you have to ask yourself – what is my competitive advantage? What am I good at? And almost inevitably that means: what do I enjoy? And the answer will go deeper than science itself. So when I look at most things that I’ve done in the lab, they’ve always had a visual component to them. In the Heidelberg screen, we identified mutants based on our abilities to look at things and recognise what was worth following up on. And to look at gastrulation is a matter of reconstructing and visualising form, and form follows force, so an understanding of physics, of flow and force and mechanics, has, to my mind, at least a strong visual component to it. I am the scientist that I am because of the talents and particular proclivities I have, and clearly painting was another expression of that same quality.
And do you still paint?
I’ve got back in to painting, but it’s challenging because I think to be really good, you have to invest more time than I have. I’m still too hung up on the idea of getting a product at the end, getting the picture to work, rather than doing a hundred versions of it to figure out how to get it to work. Every step in becoming better costs time, which of course is not very different from science, where to be really great, you have to invest and risk, and do things a bunch of times. That’s why science is not a nine to five job: so much of what we do doesn’t quite work out!
What would Development readers be surprised to find out about you?
I am an aggressive card player, and I play to win (though I must say don’t win all the time!). Trudi and I have trained all of our children to be aggressive pinochle players, and also to play to win. Pinochle is different from poker: with poker there is an element of bluff, and while I am a theatrical enough person that I could pull that off, I feel so emotionally dishonest! So I tend towards card games like pinochle where you are confronted with a problem and have to figure out how to win with the cards you have. As long as I don’t have to be dishonest about it!
Check out Development’s collection of print interviews here, and our video interview collection here


 (6 votes)
(6 votes)