January in preprints
Posted by the Node, on 3 February 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. See June’s introductory post for background, and let us know if we missed anything
2017 started where 2016 had left off, with an number of preprints covering most corners of developmental biology, plus more relevant work from related fields. Looking at the list below, it’s clear that a mix of young and established labs are using preprints. Following the trends of last year, they were predominantly found on bioRxiv, with some also on arXiv and PeerJ Preprints.
This month features regeneration in mouse digits and whole ascidians, how body axes form in gastruloids, the link between metabolism and signalling in mice, and developmental plasticity in Arabidopsis. There also were a whole bunch of fly papers, and new tools including a new ImageJ and ‘WikiGenomes’. Happy preprinting!
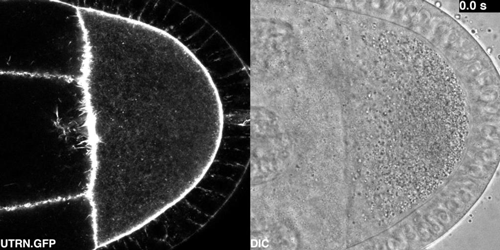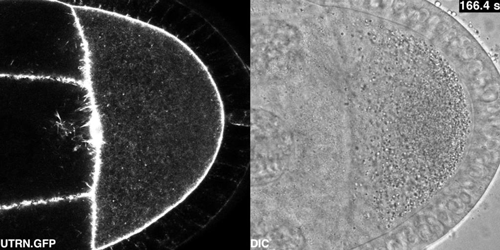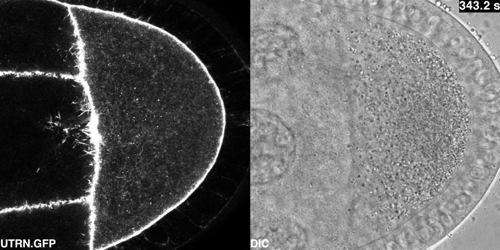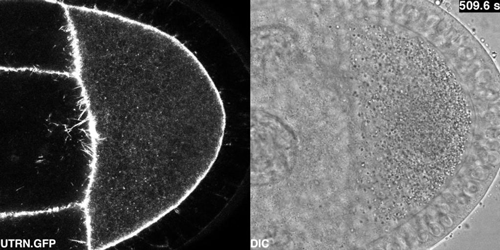
Cytoplasmic flows in the Drosophila oocyte from Drechsler, et al.
Developmental biology
| Genes & chromosomes
Abortive Initiation as a Bottleneck for Transcription in the Early Drosophila Embryo. Alexander S. Serov, Alexander J. Levine, Madhav Mani
The Drosophila Dosage Compensation Complex activates target genes by chromosome looping within the active compartment. Tamas Schauer, Yad Ghavi-Helm, Tom Sexton, Christian Albig, Catherine Regnard, Giacomo Cavalli, Eileen E M Furlong, Peter B Becker
Genome-wide Search for Zelda-like Chromatin Signatures Identifies GAF as a Pioneer Factor in Early Fly Development. Arbel Moshe, Tommy Kaplan
Polycomb-Mediated Chromatin Loops Revealed by a Sub-Kilobase Resolution Chromatin Interaction Map. Kyle P. Eagen, Erez Lieberman Aiden, Roger D. Kornberg
The Nucleosome Remodelling and Deacetylation complex restricts Mediator access to enhancers to control transcription. Maria Xenophontos, Nicola Reynolds, Sarah Gharbi, Ewan Johnstone, Jason Signolet, Robin Floyd, Meryem Ralser, Susanne Borneloev, Sabine Dietmann, Remco Loos, Paul Bertone, Brian Hendrich
Targeted degradation of CTCF decouples local insulation of chromosome domains from higher-order genomic compartmentalization. Elphege P Nora, Anton Goloborodko, Anne-Laure Valton, Johan Harmen Gibcus, Alec Uebersohn, Nezar Abdennur, Job Dekker, Leonid Mirny, Benoit Bruneau
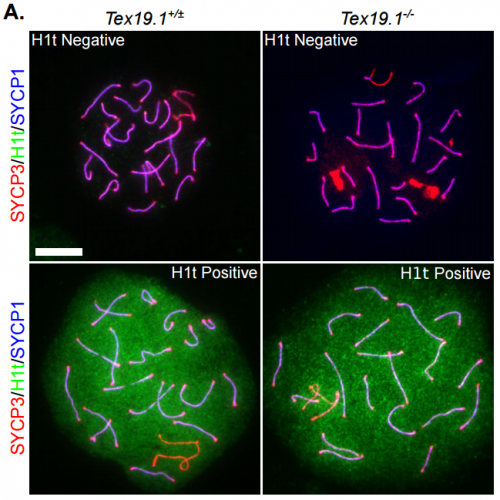
Defects in Meiotic Recombination Delay Progression Through Pachytene in Mouse Spermatocytes. James H Crichton, David R Read, Ian R Adams
Tex19.1 Regulates Meiotic DNA Double Strand Break Frequency in Mouse Spermatocytes. James H Crichton, Christopher J Playfoot, Marie MacLennan, David Read, Howard J Cooke, Ian R Adams
Spacer sequences separating transcription factor binding motifs set enhancer quality and strength. Marion Gueroult-Bellone, Kazuhiro R Nitta, Willi Kari, Edwin Jacox, Remy Beule Dauzat, Renaud Vincentelli, Carine Diarra, Ute Rothbacher, Christelle Dantec, Christian CAMBILLAU, Jacques Piette, Patrick Lemaire
Direct visualization of transcriptional activation by physical enhancer-promoter proximity. Hongtao Chen, Miki Fujioka, James B Jaynes, Thomas Gregor
Diverse yet highly selective interorgan crosstalk mechanisms shape the bodywide transcriptome landscape. Norio Takada, Madoka Omae, Fumihiko Sagawa, Neil Chi, Satsuki Endo, Satoshi Kozawa, Thomas N Sato
| Patterning & signalling
A defined platform of human peri-gastrulation-like biological fate patterning reveals coordination between Reaction-Diffusion and Positional-Information. Mukul Tewary, Joel E Ostblom, Nika Shakiba, Peter W Zandstra
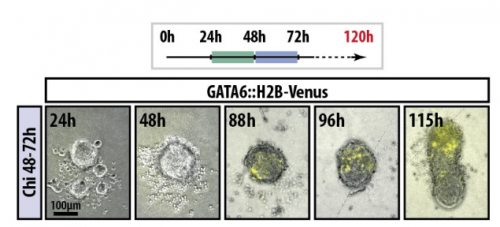
Gastruloids develop the three body axes in the absence of extraembryonic tissues and spatially localised signalling. David Turner, Luz Alonso-Crisostomo, Mehmet Girgin, Peter Baillie-Johnson, Cherise R Glodowski, Penelope C Hayward, Jérôme Collignon, Carsten Gustavsen, Palle Serup, Benjamin Steventon, Matthias Lutolf, Alfonso Martinez Arias
Early frameshift alleles of zebrafish tbx5a that fail to develop the heartstrings phenotype. Elena Chiavacci, Lucia Kirchgeorg, Anastasia Felker, Alexa Burger, Christian Mosimann
Multiple zebrafish atoh1 genes specify a diversity of neuronal types in the zebrafish cerebellum. Chelsea U. Kidwell, Chen-Ying Su, Masahiko Hibi, Cecilia B. Moens
Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. Andreas Sagner, Zachary Gaber, Julien Delile, Jennifer H Kong, David L Rousso, Caroline A Pearson, Steven E Weicksel, Neda Mousavy Gharavy, James Briscoe, Bennett Novitch
Warburg-like metabolism coordinates FGF and Wnt signaling in the vertebrate embryo. Masayuki Oginuma, Philippe Moncuquet, Fengzhu Xiong, Edward Karoly, Jerome Chal, Karine Guevorkian, Olivier Pourquie
Semaphorin-Plexin signaling influences early ventral telencephalic development and thalamocortical axon guidance. Manuela D. Mitsogiannis, Graham E. Little, Kevin J Mitchell
A damped oscillator imposes temporal order on posterior gap gene expression in Drosophila. Berta Verd, Erik Clark, Karl R. Wotton, Hilde Janssens, Eva Jimenez-Guri, Anton Crombach, Johannes Jaeger
Dynamic Maternal Gradients Control Timing and Shift-Rates for Gap Gene Expression. Berta Verd, Anton Crombach, Johannes Jaeger
Functional regulatory evolution outside of the minimal even-skipped stripe 2 enhancer. Justin Crocker, David L. Stern
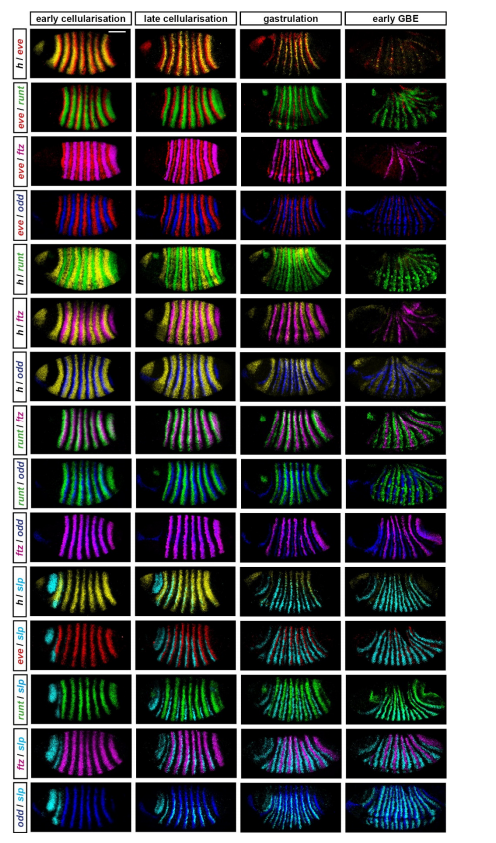
Dynamic patterning by the Drosophila pair-rule network reconciles long-germ and short-germ segmentation. Erik Clark
The scaffolding protein Cnk Interacts with Alk to Promote Visceral Founder Cell Specification in Drosophila. Georg Wolfstetter, Kathrin Pfeifer, Jesper R. vanDijk, Fredrik Hugosson, Xiangyi Lu, Ruth H. Palmer
PEAPOD limits developmental plasticity in Arabidopsis. Derek W. R. White
| Morphogenesis & Mechanics
Quantification of myosin distribution predicts global morphogenetic flow in the fly embryo. Sebastian J Streichan, Matthew F Lefebvre, Nicholas Noll, Eric F Wieschaus, Boris I Shraiman
Active diffusion and advection in the Drosophila ooplasm result from the interplay of the actin and microtubule cytoskeletons. Maik Drechsler, Fabio Giavazzi, Roberto Cerbino, Isabel M Palacios
Myosin II activity is not required for Drosophila tracheal branching morphogenesis. Amanda Ochoa-Espinosa, Stefan Harmansa, Emmanuel Caussinus, Markus Affolter
Astral microtubule dynamics regulate anaphase oscillation onset and set a robust final position of the C. elegans zygote spindle. Helene Bouvrais, Laurent Chesneau, Sylvain Pastezeur, Marie Delattre, Jacques Pecreaux
Cortical elasticity determines the geometry of prospective mesoderm constriction in Drosophila melanogaster. Konstantin Doubrovinski
| Stem cells, regeneration & ageing
Molecular and functional variation in iPSC-derived sensory neurons. Jeremy Schwartzentruber, Stefanie Foskolou, Helena Kilpinen, Julia Rodrigues, Kaur Alasoo, Andrew J Knights, Minal Patel, Angela Goncalves, Rita Ferreira, Caroline L Benn, Anna Wilbrey, Magda Bictash, Emma Impey, Lishuang Cao, Sergio Lainez, Alexandre J Loucif, Paul J Whiting, HIPSCI Consortium, Alex Gutteridge,Daniel J Gaffney
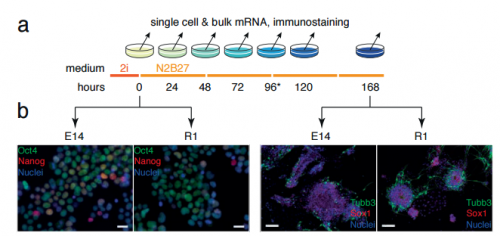
Stem cell differentiation is a stochastic process with memory. Patrick S. Stumpf, Rosanna C. G. Smith, Michael Lenz, Andreas Schuppert, Franz-Josef Müller, Ann Babtie, Thalia E. Chan, Michael P. H. Stumpf, Colin P. Please, Sam D. Howison, Fumio Arai, Ben D. MacArthur
Production of Multilayer Cell Mass from Olfactory Stem Cell and Its Utilization Potential in Nervous Tissue Regeneration. Olga Nehir Nehir Oztel, Adil M. Allahverdiyev, Aysegul Batioglu Karaaltin, Melahat Bagirova, Ercument Ovali
Non-viral induction of transient cell reprogramming in skeletal muscle to enhance tissue regeneration. Irene de Lazaro, Acelya Yilmazer, Yein Nam, Sarah Qubisi, Fazilah Maizatul Abdul Razak, Giulio Cossu, Kostas Kostarelos
Targeted gene correction of FKRP by CRISPR/Cas9 restores functional glycosylation of α-dystroglycan in cortical neurons derived from human induced pluripotent stem cells. Beatrice Lana, Jihee Kim, David Ryan, Evangelos Konstantinidis, Sandra Louzada, Beiyuan Fu, Fengtang Yang, Derek L. Stemple, Pentao Liu, Francesco Muntoni, Yung-Yao Lin
Integrated time-lapse and single-cell transcription studies highlight the variable and dynamic nature of human hematopoietic cell fate commitment. Alice Moussy, Jeremie Cosette, Romuald Parmentier, Cindy da Silva, Guillaume Corre, Angelique Richard, Olivier Gandrillon, Daniel Stockholm, Andras Paldi
Glial and stem cell expression of Fibroblast Growth Factor Receptor 1 in the embryonic and perinatal nervous system. Jantzen C Collette, Lisha Choubey, Karen Muller Smith.
Transcriptome analysis of genetically matched human induced pluripotent stem cells disomic or trisomic for chromosome 21. Patrick K. Gonzales, Virginia Fonte, Christine M. Roberts, Connor Jacobsen, Gretchen H Stein, Christopher D. Link
Yorkie is required to restrict the injury responses in planarians. Alexander Y.T. Lin, Bret J Pearson
Macrophages are required to coordinate mouse digit tip regeneration. Ken Muneoka, Jennifer Simkin, Mimi Sammarco, Luis Marrero, Lindsay Dawson, Mingquan Yan, Catherine Tucker, Alex Cammack
Histological and haematological analysis of the ascidian Botrylloides leachii (Savigny, 1816) during whole-body regeneration. Simon Blanchoud, Lisa Zondag, Miles K Lamare, Megan J Wilson
Multi-generational silencing dynamics control cell aging. Yang Li, Meng Jin, Richard O’Laughlin, Lev S Tsimring, Lorraine Pillus, Jeff Hasty, Nan Hao
Age-dependence and aging-dependence: The case of neuronal loss and lifespan in a C. elegans model of Parkinson’s disease. Javier Apfeld, Walter Fontana
The differential spatiotemporal expression pattern of shelterin genes throughout lifespan. Kay-Dietrich Wanger, Yilin Ying, Waiian Leong, Jie Jiang, Xuefei Hu, Yi Chen, Jean-Francois Michiels, Yiming Lu, Eric Gilson, Nicole Wagner, Jing Ye
Cell biology
Mapping load-bearing in the mammalian spindle reveals local kinetochore-fiber anchorage that provides mechanical isolation and redundancy. Mary Williard Elting, Manu Prakash, Dylan B. Udy, Sophie Dumont
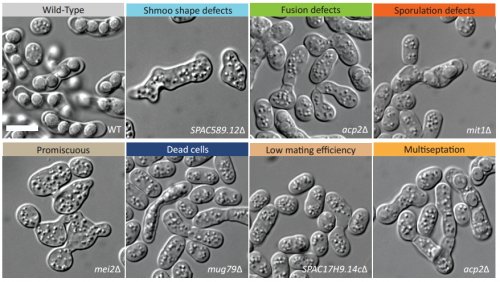
A systematic screen for morphological abnormalities during fission yeast sexual reproduction identifies a mechanism of actin aster formation for cell fusion. Omaya Dudin, Laura Merlini, Felipe O Bendezu, Raphael Groux, Vincent Vincenzetti, Sophie G Martin
Photorelease of Diacylglycerol Increases the Amplitude and Duration of Protein Kinase C-BetaII Relocation in cyto. Joachim Goedhart, Theodorus W.J. Gadella
Crawling chytrid fungi implicate actin regulators WASP and SCAR in an ancient mode of cell motility. Lillian K. Fritz-Laylin, Samuel J. Lord, R. Dyche Mullins
Distinguishing Mechanisms Underlying EMT Tristability. Dongya Jia, Mohit Kumar Jolly, Satyendra Chandra Tripathi, Petra Den Hollander, Bin Huang, Mingyang Lu, Muge Celiktas, Esmeralda Ramirez-Pena, Eshel Ben-Jacob, Jose N. Onuchic, Samir M. Hanash, Sendurai A. Mani, Herbert Levine
Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis. Marie-Cecilia Duvernoy, Thierry Mora, Maxime Ardre, Vincent Croquette, David Bensimon, Catherine Quilliet, Jean-Marc Ghigo, Martial Balland, Christophe Beloin, Sigolene Lecuyer, Nicolas Desprat
A checkpoint roadmap for the complex cell division of Apicomplexa parasites. Carmelo A Alvarez, Elena S Suvorova
Evo-devo & evo
A miRNA catalogue and ncRNA annotation of the short-living fish Nothobranchius furzeri. Mario Baumgart, Emanuel Barth, Aurora Savino, Marco Groth, Philipp Koch, Andreas Petzold, Ivan Arisi, Matthias Platzer, Manja Marz, Alessandro Cellerino

Mutation of a palmitoyltransferase ZDHHC18-like gene is responsible for the minute wing mutation in the silkworm (Bombyx mori). Ye Yu, Xiaojing Liu, Xiao Ma, Zhongjie Zhang, Na Liu, Chengxiang Hou, Muwang Li
The RAG transposon is active through the deuterostome evolution and domesticated in jawed vertebrates. Jose Ricardo Poole, Sheng Feng Huang, An Long Xu, Pontarotti Pierre
It′s okay to be green: Draft genome of the North American Bullfrog (Rana [Lithobates] catesbeiana). S Austin Hammond, René L Warren, Benjamin P Vandervalk, Erdi Kucuk, Hamza Khan, Ewan A Gibb, Pawan Pandoh, Heather Kirk, Yongjun Zhao, Martin Jones, Andrew J Mungall, Robin Coope, Stephen Pleasance, Richard A Moore, Robert A Holt, Jessica M Round, Sara Ohora, Nik Veldhoen, Caren C Helbing, Inanc Birol
Revisiting suppression of interspecies hybrid male lethality in Caenorhabditis nematodes. Lauren E Ryan, Eric S Haag
Large-scale rewiring in a yeast hybrid. Rebecca Herbst, Dana Bar-Zvi, Sharon Reikhav, Ilya Soifer, Michal Breker, Ghil Jona, Eyal Shimoni, Maya Schuldiner, Avraham Levy, Naama Barkai
No evidence for the radiation time lag model after whole genome duplications in Teleostei. Sacha Laurent, Nicolas Salamin, Marc Robinson-Rechavi
Hybrid incompatibility caused by an epiallele. Todd Blevins, Jing Wang, David Pflieger, Frédéric Pontvianne, Craig S. Pikaard
Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Shashank Gandhi, Maximilian Haeussler, Florian Razy-Krajka, Lionel Christiaen, Alberto Stolfi
Tools & Resources
Escape Excel: a tool for preventing gene symbol and accession conversion errors. Eric A Welsh, Paul A Stewart, Brent M Kuenzi
ImageJ2: ImageJ for the next generation of scientific image data. Curtis T. Rueden, Johannes Schindelin, Mark C. Hiner, Barry E. DeZonia, Alison E. Walter, Kevin W. Eliceiri
WikiGenomes: an open Web application for community consumption and curation of gene annotation data in Wikidata. Tim E Putman, Sebastien Lelong, Sebastian Burgstaller-Muelhbacher, Andra Waagmeester, Colin Diesh, Nathan Dunn, Monica Munoz-Torres, Gregory Stupp, Andrew Su, Benjamin M Good
Measuring physiological responses to stressors using a novel Hmox1 reporter mouse. Michael McMahon, Shaohong Ding, Lourdes P Acosta-Jimenez, Tania G Frangova, Colin J Henderson, C. Roland Wolf
A toolkit for tissue-specific protein degradation in C. elegans. Shaohe Wang, Ngang Heok Tang, Pablo Lara-Gonzalez, Bram Prevo, Dhanya K Cheerambathur, Andrew D Chisholm, Arshad Desai, Karen Oegema
Whole genome sequencing and assembly of a Caenorhabditis elegans genome with complex genomic rearrangements using the MinION sequencing device. John R Tyson, Nigel J O’Neil, Miten Jain, Hugh E Olsen, Philip Hieter, Terrance P Snutch
Progress Towards a Public Chemogenomic Set for Protein Kinases and a Call for Contributions. David H Drewry, Carrow I Wells, David M Andrews, Richard Angell, Hassan Al-Ali, Alison D Axtman, Stephen J Capuzzi, Jonathan M Elkins, Peter Ettmayer, Mathias Frederiksen, Opher Gileadi, Nathanael Gray, Alice Hooper, Stefan Knapp, Stefan Laufer, Ulrich Luecking, Susanne Muller, Eugene Muratov, R. Aldrin Denny, Kumar S Saikatendu, Daniel K Treiber, William J Zuercher, Timothy M Willson
The 4D Nucleome Project. Job Dekker, Andrew S Belmont, Mitchell Guttman, Victor O Leshyk, John T Lis, Stavros Lomvardas, Leonid A Mirny, Clodagh C O’Shea, Peter J Park, Bing Ren, Joan C Ritland, Jay Shendure, Sheng Zhong, The 4D Nucleome Network
Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Bogdan Budnik, Ezra Levy, Nikolai Slavov
Interactive analysis of Long-read RNA isoforms with Iso-Seq Browser. Jingyuan Hu, Prech Uapinyoying, Jeremy Goecks
Genome Graphs. Adam M Novak, Glenn Hickey, Erik Garrison, Sean Blum, Abram Connelly, Alexander Dilthey, Jordan Eizenga, M. A. Saleh Elmohamed, Sally Guthrie, André Kahles, Stephen Keenan, Jerome Kelleher, Deniz Kural, Heng Li, Michael F Lin, Karen Miga, Nancy Ouyang, Goran Rakocevic, Maciek Smuga-Otto, Alexander Wait Zaranek, Richard Durbin, Gil McVean, David Haussler, Benedict Paten
Multiplexed dynamic imaging of genomic loci in single cells by combined CRISPR imaging and DNA sequential FISH. Yodai Takei, Sheel Shah, Sho Harvey, Lei S Qi, Long Cai
CRISPR-RT: A web service for designing CRISPR-C2c2 crRNA with improved target specificity. Houxiang Zhu, Emily Richmond, Chun Liang
The 3D genome organization of Drosophila melanogaster through data integration. Qingjiao Li, Harianto Tjong, Xiao Li, Ke Gong, Xianghong Jasmine Zhou, Irene Chiolo, Frank Alber
Bayesian inference of agent-based models: a tool for studying kidney branching morphogenesis. Ben Lambert, Adam L MacLean, Alexander G Fletcher, Alexander N Coombes, Melissa H Little, Helen M Byrne
Cell fixation and preservation for droplet-based single-cell transcriptomics. Jonathan Alles, Samantha Praktiknjo, Nikos Karaiskos, Stefanie Grosswendt, Salah Ayoub, Luisa Schreyer, Anastasiya Boltengagen, Christine Kocks, Nikolaus Rajewsky
Pebbles as Dry Ice Replacement for Snap Freezing of Rodents Brains. Rene Bernard, Larissa Mosch, Ulrich Dirnagl
Accurate alignment between correlated light, cryo-light, and electron cryo-microscopy data using sample support features. Karen L Anderson, Christopher Page, Mark F Swift, Dorit Hanein, Niels Volkmann
Research practice
Data Reuse as a Prisoner’s Dilemma: the social capital of open science. Bradly Alicea
Biocuration as an undergraduate training experience: Improving the annotation of the insect vector of Citrus greening disease. Surya Saha, Prashant S Hosmani, Krystal Villalobos-Ayala, Sherry Miller, Teresa Shippy, Andrew Rosendale, Chris Cordola, Tracey Bell, Hannah Mann, Gabe DeAvila, Daniel DeAvila, Zachary Moore, Kyle Buller, Kathryn Ciolkevich, Samantha Nandyal, Robert Mahoney, Joshua Von Voorhis, Megan Dunlevy, David Farrow, David Hunter, Taylar Morgan, Kayla Shore, Victoria Guzman, Allison Izsak, Danielle E Dixon, Liliana Cano, Andrew Cridge, Shannon Johnson, Brandi L Cantarel, Stephen Richardson, Adam English, Nan Leng, Xiaolong Cao, Haobo Jiang, Chris Childers, Mei-Ju Chen, Mirella Flores, Wayne Hunter, Michelle Cilia, Lukas A Mueller, Monica Munoz-Torres, David Nelson, Monica F Poelchau, Josh Benoit, Helen Wiersma-Koch, Tom D’elia, Susan J Brown
Is Democracy the Right System? Collaborative Approaches to Building an Engaged RDM Community. Marta Teperek, Rosie Higman, Danny Kingsley
Why not…
Variation in the microbiome of the urogenital tract of koalas (Phascolarctos cinereus) with and without “wet bottom”. Alistair Raymond Legione, Jemima Amery-Gale, Michael Lynch, Leesa Haynes, James Gilkerson, Joanne Devlin, Fiona Sansom


 (No Ratings Yet)
(No Ratings Yet)
Very useful information! Thanks for sharing the links.