June in preprints
Posted by the Node, on 5 July 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. See last year’s introductory post for background, and let us know if we missed anything
We started this feature a year ago, and each month it’s taken a little longer to curate as more labs decide to deposit preprints alongside conventional channels of publication, and as this practise is encouraged by an increasing number of journals, funding bodies and academic organisations.
Highlights this anniversary month include a lot of neural development, analyses of a controversial CRISPR off target paper (and a word from the authors of the original paper), an analysis of stem cells and mutation in long lived trees, a slew of new genomes (gar, greyling, reindeer, zucchini and colonial tunicate!), and the routine acronym-fest of new data analysis tools (incorporating mimosas and pineapples this time). The preprints were hosted on bioRxiv, PeerJ and arXiv.
Use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
| Imaging etc.
Research practice
Why not…
Developmental biology
| Patterning & signalling
Epidermal Wnt signalling regulates transcriptome heterogeneity and proliferative fate in neighbouring cells. Arsham Ghahramani, Giacomo Donati, Nicholas M. Luscombe, Fiona M. Watt
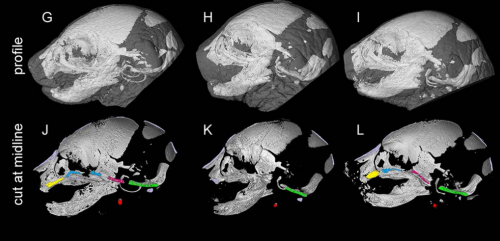
Sonic hedgehog is required for neural crest-dependent patterning of the intrinsic tongue musculature. Shigeru Okuhara, Anahid Ahmadi Birjandi, Hadeel Adel Al-Lami, Tomoko Sagai, Takanori Amano, Toshihiko Shiroishi, Karen J. Liu, Martyn Cobourne, Sachiko Iseki
Hippo signaling restricts cells in the second heart field that differentiate into Islet-1-positive atrial cardiomyocytes. Hajime Fukui, Takahiro Miyazaki, Hiroyuki Ishikawa, Hiroyuki Nakajima, Naoki Mochizuki
ICM conversion to epiblast by FGF/ERK inhibition is limited in time and requires transcription and protein degradation. Sylvain Bessonnard, Sabrina Coqueran, Sandrine Vandormael-Pournin, Alexandre Dufour, Jerome Artus, Michel Cohen-Tannoudji
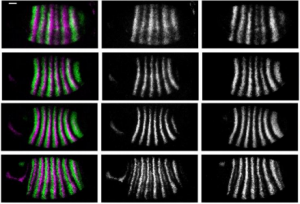
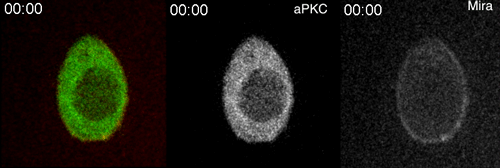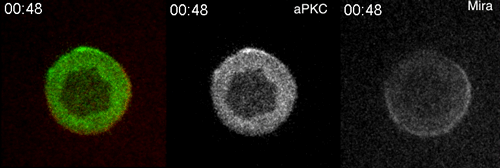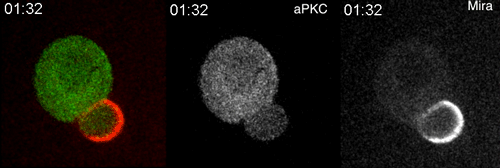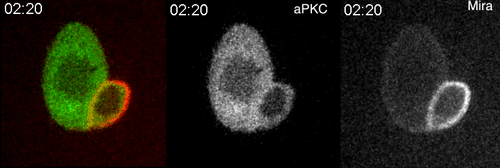
aPKC-mediated displacement and actomyosin-mediated retention polarize Miranda in Drosophila neuroblasts. Matthew Hannaford, Anne Ramat, Nicolas Loyer, Jens Januschke
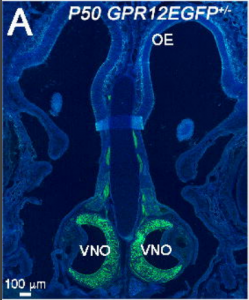
Olfactory and vomeronasal innervation of the olfactory bulbs are not essential for GnRH-1 neuronal migration to the brain. Ed Zandro M. Taroc, Aparna Prasad, Jennifer M. Lin, Paolo Forni
Insm1a regulates motor neuron development in zebrafish. Jie Gong, Xin Wang, Chenwen Zhu, Xiaohua Dong, Qinxin Zhang, Xiaoning Wang, Xuchu Duan, Fuping Qian, Yu Gao, Qingshun Zhao, Renjie Chai, Dong Liu
RAPGEF5 regulates nuclear transport of β-catenin. John Noel Griffin, Florencia del Viso, Anna R. Duncan, Andrew Robson, Saurabh Kulkarni, Karen J. Liu, Mustafa K. Khokha
Differing strategies used by motor neurons and glia to achieve robust development of an adult neuropil in Drosophila. Jonathan Enriquez, Laura Quintana Rio, Richard Blazeski, Carol Mason, Richard S. Mann
Cell type-specific expression profiling sheds light on the development of a peculiar neuron, housing a complex organelle. Kartik Sunagar, Yaara Columbus-Shenkar, Arie Fridrich, Nadya Gutkovitch, Reuven Aharoni, Yehu Moran
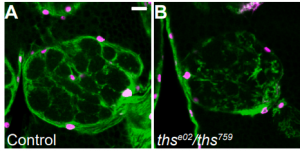
Fibroblast Growth Factor Signaling Instructs Ensheathing Glia Wrapping of Drosophila Olfactory Glomeruli. Bing Wu, Jiefu Li, Ya-Hui Chou, David Luginbuhl, Liqun Luo
Syndecan functions to regulate Wnt-dependent axon guidance in C. elegans. Samantha N Hartin, Meagan E Kurland, Brian D Ackley
Xbra and Smad-1 response elements cooperate in PV.1 promoter to inhibit the early neurogenesis in Xenopus embryos. Shiv Kumar, Zobia Umair, Jaeho Yoon, Unjoo Lee, SungChan Kim, Jae-Bong Park, Jae-Yong Lee, Jaebong Kim
Interphase-Arrested Drosophila Embryos Initiate Mid-Blastula Transition At A Low Nuclear-Cytoplasmic Ratio. Isaac Strong, Kai Yuan, Patrick H. O’Farrell
The Emergent Connectome in Caenorhabditis elegans Embryogenesis. Bradly J. Alicea
Pericyte ontogeny: the use of chimeras to track a cell lineage of diverse germ line origins. Heather Corbett Etchevers
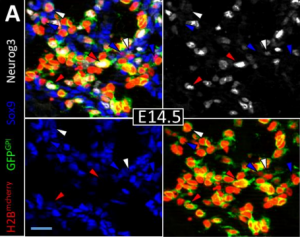
FUCCI tracking shows that Neurog3 levels vary with cell-cycle phase in endocrine-biased pancreatic progenitors. Matthew Edward Bechard, Eric D. Bankaitis, Alessandro Ustione, David Piston, Mark A. Magnuson, Christopher V. E. Wright
| Morphogenesis & mechanics
Multiple conserved cell adhesion protein interactions mediate neural wiring of a sensory circuit in C. elegans. Byunghyuk Kim, Scott Emmons
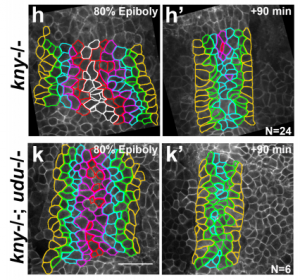
The chromatin factor Gon4l regulates embryonic axis extension by promoting mediolateral cell polarity and notochord boundary formation through negative regulation of cell adhesion. Margot L. K. Williams, Atsushi Sawada, Terin Budine, Chunyue Yin, Paul Gontarz, Lilianna Solnica-Krezel
WDR5 stabilizes actin architecture to promote multiciliated cell formation. Saurabh S. Kulkarni, John N. Griffin, Karel F. Liem, Mustafa K. Khokha
Distinct effects of tubulin isotype mutations on neurite growth in Caenorhabditis elegans. Chaogu Zheng, Margarete Diaz-Cuadros, Susan Laura Jao, Ken Nguyen, David H Hall, Martin Chalfie
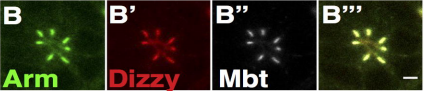
Rap1, AF6 and Pak4 cooperate with Par3 to promote ZA assembly and remodeling in the fly photoreceptor. Rhian F Walther, Mubarik Burki, Noelia Pinal Seoane, Clare Rogerson, Franck Pichaud
| Genes & genomes
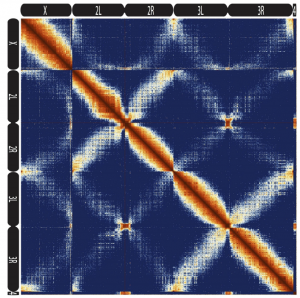
Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogaster embryo. Michael R. Stadler, Jenna E. Haines, Michael B. Eisen
Conserved noncoding transcription and core promoter regulatory code in early Drosophila development. Philippe J. Batut, Thomas R. Gingeras
A single cell transcriptional roadmap for cardiopharyngeal fate diversification. Wei Wang, Xiang Niu, Estelle Jullian, Robert G. Kelly, Rahul Satija, Lionel Christiaen
Dual functions of Discoidin domain receptor coordinate cell-matrix adhesion and collective polarity in migratory cardiopharyngeal progenitors. Yelena Y. Bernadskaya, Saahil Brahmbhatt, Stephanie E. Gline, Wei Wang, Lionel Christiaen
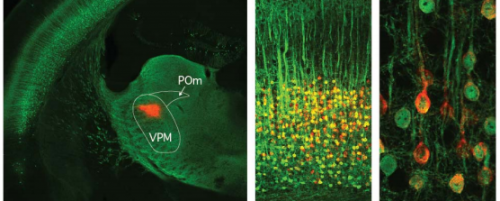
Variation in neuronal activity state, axonal projection target, and position principally define the transcriptional identity of individual neocortical projection neurons. Maxime Chevée, Johanna D. Robertson, Gabrielle H. Cannon, Solange P. Brown, Loyal A. Goff
Chromatin accessibility dynamics of myogenesis at single cell resolution. Hannah Pliner, Jonathan Packer, Jose McFaline-Figueroa, Darren Cusanovich, Riza Daza, Sanjay Srivatsan, Xiaojie Qiu, Dana Jackson, Anna Minkina, Andrew Adey, Frank Steemers, Jay Shendure, Cole Trapnell
Transcript Isoform Differences Across Human Tissues Are Predominantly Driven By Alternative Start And Termination Sites Of Transcription. Alejandro Reyes, Wolfgang Huber
Genome Architecture Leads a Bifurcation in Cell Identity. Sijia Liu, Haiming Chen, Scott Ronquist, Laura Seaman, Nicholas Ceglia, Walter Meixner, Lindsey A. Muir, Pin-Yu Chen, Gerald Higgins, Pierre Baldi, Steve Smale, Alfred Hero, Indika Rajapakse
Double Maternal Effect: Duplicated Nucleoplasmin 2 Genes, npm2a And npm2b, Are Shared By Fish And Tetrapods, And Have Distinct And Essential Roles In Early Embryogenesis. Caroline T. Cheung, Jeremy Pasquier, Aurelien Bouleau, Thao-Vi Nguyen, Franck Chesnel, Yann Guiguen, Julien Bobe
Epigenetic Memory via Concordant DNA Methylation Persists in Mammalian Toti- and Pluripotent Stem Cells. Minseung Choi, Diane P Genereux, Jamie Goodson, Haneen Al-Azzawi, Shannon Q. Allain, Noah Simon, Stan Palasek, Carol B. Ware, Chris Cavanaugh, Daniel G. Miller, Winslow C. Johnson, Kevin D. Sinclair, Reinhard Stöger, Charles D. Laird
PARP mediated chromatin unfolding is coupled to long-range enhancer activation. Nezha S. Benabdallah, Iain Williamson, Robert S. Illingworth, Shelagh Boyle, Graeme R. Grimes, Pierre Therizols, Wendy Bickmore
A functional role for the epigenetic regulator ING1 in activity-induced gene expression in primary cortical neurons. Laura Leighton, Qiongyi Zhao, Xiang Li, Chuanyang Dai, Paul Robert Marshall, Sha Liu, Yi Wang, Esmi Lau Zajaczkowski, Nitin Khandelwal, Arvind Kumar, Timothy William Bredy, Wei Wei
Molecular analysis of the midbrain dopaminergic niche during neurogenesis. Enrique M. Toledo, Gioele La Manno, Pia Rivetti di Val Cervo, Daniel Gyllborg, Saiful Islam, Carlos Villaescusa, Sten Linnarsson, Ernest Arenas
The Drosophila Y chromosome affects heterochromatin integrity genome-wide. Emily Brown, Doris Bachtrog
Mating can cause transgenerational gene silencing in Caenorhabditis elegans. Sindhuja Devanapally, Samual Allgood, Antony M. Jose
| Stem cells, regeneration & disease modelling
Epigenetic resetting of human pluripotency. Ge Guo, Ferdinand von Meyenn, Maria Rostovskaya, James Clarke, Sabine Dietmann, Duncan Baker, Anna Sahakyan, Samuel Myers, Paul Bertone, Wolf Reik, Kathrin Plath, Austin Smith
In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Alyssa J Miller, Melinda S Nagy, David R Hill, Yu-Hwai Tsai, Yoshiro Aoki, Briana R Dye, Alana M Chin, Sha Huang, Michael A.H. Ferguson, Felix Zhou, Eric S. White, Vibha Lama, Jason R. Spence
Epidermal stem cells self-renew upon neighboring differentiation. Kailin R. Mesa, Kyogo Kawaguchi, David G. Gonzalez, Katie Cockburn, Jonathan Boucher, Tianchi Xin, Allon M. Klein, Valentina Greco
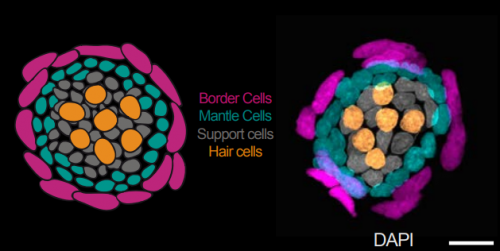
Neural stem cells induce the formation of their physical niche during organogenesis. Ali Seleit, Isabel Krämer, Bea Riebesehl, Elizabeth Mayela Ambrosio, Julian Stolper, Nicolas Dross, Colin Lischik, Lazaro Centanin
Inter-organ regulation of Drosophila intestinal stem cell proliferation by a hybrid organ boundary zone. Jessica Sawyer, Erez Cohen, Donald Fox
Low Rate of Somatic Mutations in a Long-Lived Oak Tree. Namrata Sarkar, Emanuel Schmid-Siegert, Christian Iseli, Sandra Calderon, Caroline Gouhier-Darimont, Jacqueline Chrast, Pietro Cattaneo, Frederic Schutz, Laurent Farinelli, Marco Pagni, Michel Schneider, Jeremie Voumard, Michel Jaboyedoff, Christian Fankhauser, Christian S. Hardtke, Laurent Keller, John R. Pannell, Alexandre Reymond, Marc Robinson-Rechavi, Ioannis Xenarios, Philippe Reymond
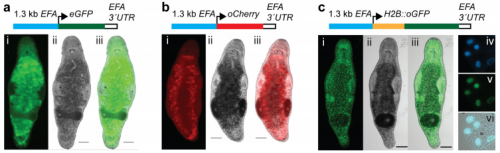
A platform for efficient transgenesis in Macrostomum lignano, a flatworm model organism for stem cell research. Jakub Wudarski, Daniil Simanov, Kirill Ustyantsev, Katrien de Mulder, Margriet Grelling, Magda Grudniewska, Frank Beltman, Lisa Glazenburg, Turan Demircan, Julia Wunderer, Weihong Qi, Dita B. Vizoso, Philipp M. Weissert, Daniel Olivieri, Stijn Mouton, Victor Guryev, Aziz Aboobaker, Lukas Scharer, Peter Ladurner, Eugene Berezikov
CFTR Modulates Wnt/β-Catenin Signaling and Stem Cell Proliferation in Murine Intestine. Ashlee M Strubberg, Jinghua Liu, Nancy M Walker, Casey D Stefanski, R John MacLeod, Scott T Magness, Lane L Clarke
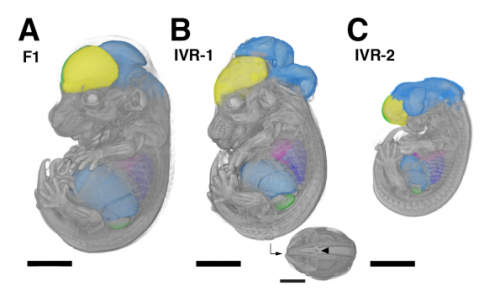
Genetic mapping of species differences via “in vitro crosses” in mouse embryonic stem cells. Stefano Lazzarano, Marek Kučka, João P. L. Castro, Ronald Naumann, Paloma Medina, Michael N. C. Fletcher, Rebecka Wombacher, Joost Gribnau, Tino Hochepied, Claude Libert, Yingguang Frank Chan
Replacing reprogramming factors with antibodies selected from autocrine antibody libraries. Joel W. Blanchard, Jia Xie, Nadja El-Mecharrafie, Simon Gross, Sohyon Lee, Richard Lerner, Kristin K. Baldwin
Widespread translational remodeling during human neuronal differentiation. John D Blair, Dirk Hockemeyer, Jennifer A Doudna, Helen S Bateup, Stephen N Floor
Automatic identification of informative regions with epigenomic changes associated to hematopoiesis. Enrique Carrillo de Santa Pau, David Juan, Vera Pancaldi, Felipe Were, Jose Ignacio Martin-Subero, Daniel Rico, Alfonso Valencia
Human-specific genomic features of pluripotency regulatory networks link NANOG with fetal and adult brain development. Gennadi Glinsky
Caspase-8, RIPK1, and RIPK3 Coordinately Regulate Retinoic Acid-Induced Cell Differentiation and Necroptosis. Masataka Someda, Shunsuke Kuroki, Makoto Tachibana, Shin Yonehara
A Positive Feedback Loop Ensures Propagation of ROS Production and JNK Signaling Throughout Drosophila Tissue Regeneration. Sumbul Jawed Khan, Syeda Nayab Fatima Abidi, Andrea Skinner, Yuan Tian, Rachel K. Smith-Bolton
Post-transcriptional modulation of Dscam1 enhances axonal growth in development and after injury. Marta Koch, Maya Nicolas, Marlen Zschaetzsch, Natalie de Geest, Annelies Claeys, Jiekun Yan, Matthew Morgan, Marie-Luise Erfurth, Matthew Holt, Dietmar Schmucker, Bassem Hassan
Sex differences in gene regulation in the dorsal root ganglion after nerve injury. Kimberly E. Stephens, Weiqiang Zhou, Zhicheng Ji, Shaoqiu He, Hongkai Ji, Yun Guan, Sean D. Taverna
Obesity/Type II Diabetes Alters Macrophage Polarization Resulting in a Fibrotic Tendon Healing Response. Jessica E Ackerman, Michael B Geary, Caitlin A Orner, Fatima Bawany, Alayna Loiselle
Deletion Of EP4 In S100a4-Lineage Cells Reduces Scar Tissue Formation During Early But Not Later Stages Of Tendon Healing. Jessica E. Ackerman, Katherine T. Best, Regis J. O’Keefe, Alayna Loiselle
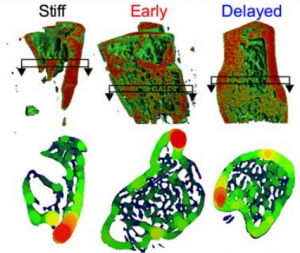
Recapitulating bone development for tissue regeneration through engineered mesenchymal condensations and mechanical cues. Anna M. McDermott, Samuel Herberg, Devon E. Mason, Hope B. Pearson, James H. Dawahare, Joseph M. Collins, Mark W. Grinstaff, Daniel J. Kelly, Eben Alsberg, Joel D. Boerckel
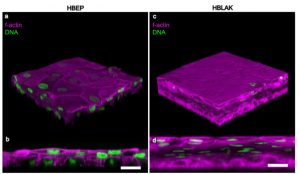
A urine-dependent human urothelial organoid offers a viable alternative to rodent models of infection. Harry Horsley, Dhanuson Dharmasena, James Malone-Lee, Jennifer L. Rohn
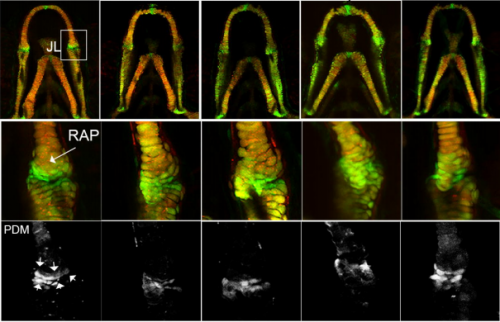
A zebrafish model of developmental joint dysplasia: Manipulating the larval mechanical environment to drive the malformation and recovery of joint shape. Karen A. Roddy, Roddy E. H. Skinner, Lucy H. Brunt, Erika Kague, Stephen Cross, Emily J. Rayfield, Chrissy L. Hammond
FOXS1 is a Master Regulator of Pathological Epithelial to Mesenchymal Transition in Human Epithelia. Timothy A Blenkinsop, Thomasz Swigut, Nathan Boles, Rajini Srinivasan, Alvaro Rada-Iglesias, Qingjie Wang, Jeffrey Stern, Joanna Wysocka, Sally Temple
A comprehensive portrait of cilia and ciliopathies from a CRISPR-based screen for Hedgehog signaling. David K. Breslow, Sascha Hoogendoorn, Adam R. Kopp, David W. Morgens, Brandon K. Vu, Kyuho Han, Amy Li, Gaelen T. Hess, Michael C. Bassik, James K. Chen, Maxence V. Nachury
Mutations in the Drosophila splicing regulator Prp31 as a model for Retinitis pigmentosa 11. Malte Lehmann, Sarita Hebbar, Holger Brandl, Weihua Leng, Naharajan Lakshmanaperumal, Sylke Winkler, Elisabeth Knust
Effects of Prenatal Stress on Structural Brain Development and Aging in Humans. Katja Franke, Bea van den Bergh, Susanne R. de Rooij, Tessa J. Roseboom, Peter W. Nathanielsz, Otto W. Witte, Matthias Schwab
Chd8 haploinsufficient mice display anomalous behaviours, increased brain size and cortical hyper-connectivity. Philipp Suetterlin, Shaun Hurley, Conor Mohan, Kimberley L. H. Riegman, Marco Pagani, Angela Caruso, Jacob Ellegood, Alberto Galbusera, Ivan Crespo-Enriquez, Caterina Michetti, Robert Ellingford, Olivier Brock, Alessio Delogu, Philippa Francis-West, Jason P. Lerch, Maria Luisa Scattoni, Alessandro Gozzi, Cathy Fernandes, Albert Basson
Homeodomain interacting protein kinase promotes tumorigenesis and metastatic cell behavior. Jessica A. Blaquiere, Kenneth Kin Lam Wong, Stephen D. Kinsey, Jin Wu, Esther M. Verheyen
Evo-devo & evo
Characterization Of NvLWamide-Like Neurons Reveals Stereotypy In Nematostella Nerve Net Development. Jamie Havrilak, Dylan Faltine-Gonzalez, Yiling Wen, Daniella Fodera, Ayanna Simpson, Craig Magie, Michael J. Layden
wingless is a positive regulator of eyespot color patterns in Bicyclus anynana butterflies. Nesibe Ozsu, Qian Yi Chan, Bin Chen, Mainak Das Gupta, Antonia Monteiro

Melanism patches up the defective cuticular morphological traits through promoting the up-regulation of cuticular protein-coding genes in Bombyx mori. Liang Qiao, Ri-xin Wang, You-jin Hao, Hai Hu, Gao Xiong, Song-zhen He, Jiang-bo Song, Kun-peng Lu,James Mallet, Ya-qun Xin, Bin Chen, Fang-yin Dai
High-quality genome assemblies uncover caste-specific long non-coding RNAs in ants. Emily J. Shields, Roberto Bonasio
Unravelling the genomic basis and evolution of the pea aphid male wing dimorphism. Binshuang Li, Ryan D. Bickel, Benjamin J. Parker, Neetha N. Vellichirammal, Mary Grantham, Jean-Christophe Simon, David L. Stern, Jennifer A. Brisson
Hydra, a model system for deciphering the mechanisms of aging and resistance to aging. Quentin Schenkelaars, Szymon Tomczyk, Yvan Wenger, Kazadi Ekundayo, Victor Girard, Wanda Buzgariu, Steve Austad, Brigitte Galliot
Human-specific changes in two functional enhancers of FOXP2. Antonio Benitez-Burraco, Raul Torres-Ruiz, Pere Gelabert Xirinachs, Carles Lalueza-Fox, Sandra Rodriguez-Perales, Paloma Garcia-Bellido
Punctuated evolution shaped modern vertebrate diversity. Michael Landis, Joshua G. Schraiber
Selective constraints on coding sequences of nervous system genes are a major determinant of duplicate gene retention in vertebrates. Julien Roux, Jialin Liu, Marc Robinson-Rechavi
Patterns of divergence across the geographic and genomic landscape of a butterfly hybrid zone associated with a climatic gradient. Sean Ryan, Michael C. Fontaine, J. Mark Scriber, Michael E. Pfrender, Shawn T. O’Neil, Jessica J. Hellmann
De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution. Simon Blanchoud, Kim Rutherford, Lisa Zondag, Neil Gemmell, Megan J. Wilson
Evolution of gene expression after whole-genome duplication: new insights from the spotted gar genome. Jeremy Pasquier, Ingo Braasch, Peter Batzel, Cedric Cabau, Jerome Montfort, Thaovi Nguyen, Elodie Jouanno, Camille Berthelot, Christophe Klopp, Laurent Journot, John Postlethwait, Yann Guiguen, Julien Bobe
The grayling genome reveals selection on gene expression regulation after whole genome duplication. Srinidhi Varadharajan, Simen R. Sandve, Ole K. Tørresen, Sigbjørn Lien, Leif Asbjørn Vollestad, Sissel Jentoft, Alexander J. Nederbragt, Kjetill S. Jakobsen
Draft genome of the Reindeer (Rangifer tarandus). Zhipeng Li, Zeshan Lin, Lei Chen, Hengxing Ba, Yongzhi Yang, Kun Wang, Wen Wang, Qiang Qiu, Guangyu Li
De-novo assembly of zucchini genome reveals a whole genome duplication associated with the origin of the Cucurbita genus. Javier Montero-Pau, eJosé Blanca, Aureliano Bombarely, Pello Ziarsolo, Cristina Esteras, Carlos Martí-Gómez, María Ferriol, Pedro Gómez, Manuel Jamilena, Lukas Mueller, Belén Picó, Joaquín Cañizares
A dead gene walking: convergent degeneration of a clade of MADS-box genes in Brassicaceae. Andrea Hoffmeier, Lydia Gramzow, Amey S. Bhide, Nina Kottenhagen, Andreas Greifenstein, Olesia Schubert, Klaus Mummenhoff, Annette Becker, Guenter Theissen
Evidence for “inter- and intraspecific horizontal genetic transfers” between anciently asexual bdelloid rotifers is explained by cross-contamination. Christopher G Wilson, Reuben W Nowell, Timothy G Barraclough
Fine scale mapping of genomic introgressions within the Drosophila yakuba clade. David A. Turissini, Daniel R. Matute
Evolutionary proteomics uncovers ciliary signaling components. Monika Abedin Sigg, Tabea Menchen, Jeffery Johnson, Chanjae Lee, Semil P. Choksi, Galo Garcia III, Henriette Busengdal, Gerard Dougherty, Petra Pennekamp, Claudius Werner, Fabian Rentzsch, Nevan Krogan, John B. Wallingford, Heymut Omran, Jeremy F. Reiter
Cell biology
The β3-integrin endothelial adhesome regulates microtubule dependent cell migration. Samuel J. Atkinson, Aleksander M. Gontarczyk, Tim S. Ellison, Robert T. Johnson, Benjamin M. Kirkup, Abdullah Alghamdi, Wesley J. Fowler, Bernardo C. Silva, Jochen J. Schneider, Katherine N. Weilbaecher, Mette M. Mogensen, Mark D. Bass, Dylan R. Edwards, Stephen D. Robinson
An adder behavior in mammalian cells achieves size control by modulation of growth rate and cell cycle duration. Clotilde Cadart, Sylvain Monnier, Jacopo Grilli, Rafaele Attia, Emmanuel Terriac, Buzz Baum, Marco Cosentino-Lagomarsino, Matthieu Piel
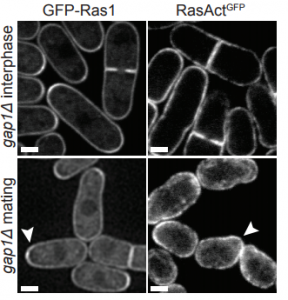
Feedback inhibition of Ras activity coordinates cell fusion with cell-cell contact. Laura Merlini, Bita Khalili, Omaya Dudin, Laetitia Michon, Vincent Vincenzetti, Sophie G Martin
Proteomics of phosphorylation and protein dynamics during fertilization and meiotic exit in the Xenopus egg. Marc Stuart Presler, Elizabeth Van Itallie, Allon M. Klein, Ryan Kunz, Peg Coughlin, Leonid Peshkin, Steven Gygi, Martin Wühr, Marc Kirschner
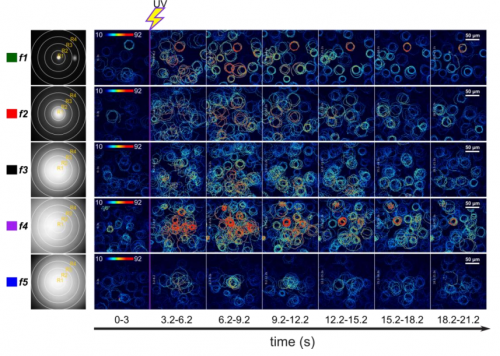
Sperm chemotaxis is driven by the slope of the chemoattractant concentration field. Héctor Vicente Ramírez-Gómez, Vilma Jiménez-Sabinina, Idan Tuval, Martín Velázquez-Pérez, Carmen Beltrán, Jorge Carneiro, Christopher Wood, Alberto Darszon, VAdán Guerrero
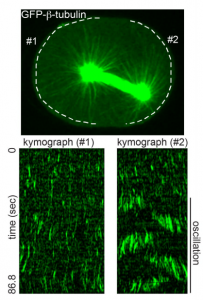
The tumor suppressor APC is an attenuator of spindle-pulling forces during C. elegans asymmetric cell division. Kenji Sugioka, Lars-Eric Fielmich, Kota Mizumoto, Bruce Bowerman, Sander van den Heuvel, Akatsuki Kimura, Hitoshi Sawa
Intrinsically disordered regions contribute promiscuous interactions to RNP granule assembly. David S. W. Protter, Bhalchandra S. Rao, Briana Van Treeck, Yuan Lin, Laura Mizoue, Michael K. Rosen, Roy Parker
A First Order Phase Transition Underlies the Formation of Sub-Diffractive Protein Aggregates in Mammalian Cells. Arjun Narayanan,Anatoli B. Meriin, Michael Y. Sherman, Ibrahim I. Cisse
Endocytosis caused by liquid-liquid phase separation of proteins. Louis-Philippe Bergeron-Sandoval, Hossein Khadivi Heris, Adam G. Hendricks, Allen J. Ehrlicher, Paul Francois, Rohit V. Pappu, Stephen W. Michnick
Principles that govern competition or co-existence in Rho-GTPase driven polarization. Jian-geng Chiou, Timothy C. Elston, Thomas P. Witelski, David G. Schaeffer, Daniel J. Lew
Crosslinkers both drive and brake cytoskeletal remodeling and furrowing in cytokinesis. Carlos Patino Descovich, Daniel B Cortes, Sean Ryan, Jazmine Nash, Li Zhang, Paul S Maddox, Francois Nedelec, Amy Shaub Maddox
Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. Jennifer T. Wang, Dong Kong, Christian R. Hoerner, Jadranka Loncarek,T im Stearns
The Centrosome Controls the Number and Spatial Distribution of Microtubules by Negatively Regulating Alternative MTOCs. Maria P. Gavilan, Pablo Gandolfo, Fernando R. Balestra, Francisco Arias, Michel Bornens, Rosa M. Rios
Lis1 has two opposing modes of regulating cytoplasmic dynein. Morgan E. DeSantis, Michael A. Cianfrocco, Zaw M. Htet, Phuoc T. Tran, Samara L. Reck-Peterson, Andres E. Leschziner
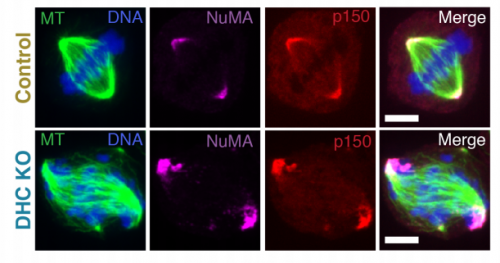
NuMA Targets Dynein to Microtubule Minus-Ends at Mitosis. Christina Hueschen, Samuel J. Kenny, Ke Xu, Sophie Dumont
Dynamics of the IFT Machinery at the Ciliary Tip. Alexander Chien, Sheng Min Shih, Raqual Bower, Douglas Tritschler, Mary E. Porter, Ahmet Yildiz
TRIM9-dependent ubiquitination of DCC constrains kinase signaling, exocytosis, and axon branching. Melissa Plooster, Shalini Menon, Cortney C. Winkle, Fabio L. Urbina, Caroline Monkiewicz, Kristen D. Phend, Richard J. Weinberg, Stephanie L. Gupton
Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. David Kern, Elizabeth Wilson-Kubalek, Iain Cheeseman
Akt/PKB enhances non-canonical Wnt signals by compartmentalizing β-Catenin. Nicolas Aznar, Nina Sun, Ying Dunkel, Jason Ear, Matthew Buschman, Pradipta Ghosh
Cargo crowding at actin-rich regions along axons causes local traffic jams in neurons. Parul Sood, Kausalya Murthy, T. Vinod Kumar, Michael L. Nonet, Gautam I. Menon, Sandhya P. Koushika
The Drosophila Formin Fhod Nucleates Actin Filaments. Aanand A Patel, Zeynep A Oztug Durer, Aaron P van Loon, Margot E Quinlan
Cofilin Drives Rapid Turnover and Fluidization of Entangled F-actin. Patrick M. McCall, Frederick C. MacKintosh, David R. Kovar, Margaret L. Gardel
The Neuron Specific Formin Delphilin Nucleates Actin Filaments but Does Not Enhance Elongation. William T. Silkworth, Kristina L. Kunes, Grace C. Nickel, Martin L. Phillips, Margot E. Quinlan, Christina L. Vizcarra
EFA6 regulates selective polarised transport and axon regeneration from the axon initial segment. Richard Eva, Hiroaki Koseki, Venkateswarlu Kanamarlapudi, James Fawcett
Tension-dependent stretching and folding of ZO-1 controls the localization of its interactors. Domenica Spadaro, Shimin Le, Thierry Laroche, Isabelle Mean, Lionel Jond, Jie Yan, Sandra Citi
Meiotic Crossover Patterning In The Absence Of ATR: Loss Of Interference And Assurance But Not The Centromere Effect. Morgan M Brady, Susan McMahan, Jeff Sekelsky
Linearly changing stress environment causes cellular growth phenotype. Guoliang Li, Benjamin K. Kesler, Alexander Thiemicke, Dustin C. Rogers, Gregor Neuert
Cell code: the evolving arrangement of molecules that perpetuates life. Antony M. Jose
Modelling
Synthesizing developmental trajectories. Paul Villoutreix, Joakim Andén, Bomyi Lim, Hang Lu, Ioannis G. Kevrekidis, Amit Singer, Stanislav Y. Shvartsman
Adhesion and volume constraints via nonlocal interactions lead to cell sorting. J. A. Carrillo, A. Colombi, M. Scianna
A geometrically controlled rigidity transition in a model for confluent 3D tissues. Matthias Merkel, Lisa Manning
Scalable composition frameworks for multicellular logic. Sarah Guiziou, Pauline Mayonove, Federico Ulliana, Violaine Moreau, Michel Leclere, Jerome Bonnet
A novel mathematical method for disclosing oscillations in gene transcription: a comparative study. Athanasios C. Antoulas, Bokai Zhu, Qiang Zhang, Brian York,Bert W. O’Malley, Clifford Dacso
Tools & resources
| Imaging etc.
BPG: Seamless, Automated and Interactive Visualization of Scientific Data. Christine P’ng, Jeffrey Green, Lauren C. Chong, Daryl Waggott, Stephenie D. Prokopec, Mehrdad Shamsi, Francis Nguyen, Denise Y. F. Mak, Felix Lam, Marco A. Albuquerque, Ying Wu, Esther H. Jung, Maud H. W. Starmans, Michelle A. Chan-Seng-Yue, Cindy Q. Yao, Bianca Liang, Emilie Lalonde, Syed Haider, Nicole A. Simone, Dorota Sendorek, Kenneth C. Chu, Nathalie C. Moon, Natalie S. Fox, Michal R. Grzadkowski, Nicholas J. Harding, Clement Fung, Amanda R. Murdoch, Kathleen E. Houlahan, Jianxin Wang, David R. Garcia, Richard de Borja, Ren X. Sun, Xihui Lin, Gregory M. Chen, Aileen Lu, Yu-Jia Shiah, Amin Zia, Ryan Kearns, Paul Boutros
Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Pornchai Kaewsapsak, David Michael Shechner, William Mallard, John L. Rinn, Alice Y. Ting
ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. Valentina De Col, Philippe Fuchs, Thomas Nietzel, Marlene Elsaesser, Chia Pao Voon, Alessia Candeo, Ingo Seeliger, Mark Fricker, Christopher Grefen, Ian Max Moller, Andrea Bassi, Boon Leong Lim, Marco Zancani, Andreas Meyer, Alex Costa, Stephan Wagner, Markus Schwarzlaender
Multiplexed Live-Cell Visualization Of Endogenous Proteins With Nanometer Precision By Fluorobodies. Alina Klein, Susanne Hank, Anika Raulf, Felicitas Tissen, Mike Heilemann, Ralph Wieneke, Robert Tampé
Impact of fluorescent protein fusions on the bacterial flagellar motor. Minyoung Heo, Ashley L. Nord, Delphine Chamousset, Erwin van Rijn, Bertus J. E. Beaumont, Francesco Pedaci
Matryoshka: Ratiometric biosensors from a nested 1 cassette of green- and orange-emitting fluorescent proteins. Cindy Ast, Jessica Foret, Luke Oltrogge, Roberto De Michele, Thomas Kleist, Cheng-Hsun Ho, Wolf Frommer
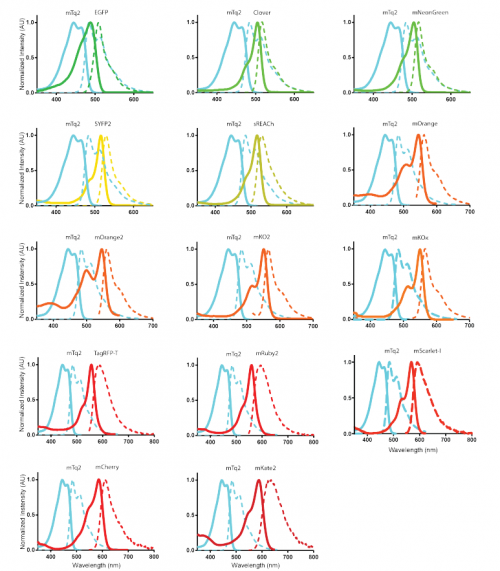
Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2. Marieke Mastop, Daphne S. Bindels, Nathan C. Shaner, Marten Postma, Theodorus W. J. Gadella Jr., Joachim Goedhart
48-spot single-molecule FRET setup with periodic acceptor excitation. Antonino Ingargiola, Maya Segal, Angelo Gulinatti, Ivan Rech, Ivan Labanca, Piera Maccagnani, Massimo Ghioni, Shimon Weiss, Xavier Michalet
A transgenic toolkit for visualizing and perturbing microtubules reveals unexpected functions in the epidermis. Andrew Muroyama, Terry Lechler
An optimized pipeline for parallel image-based quantification of gene expression and genotyping after in situ hybridization. Tomasz Dobrzycki, Monika Krecsmarik, Florian Bonkhofer, Roger K Patient, Rui Monteiro
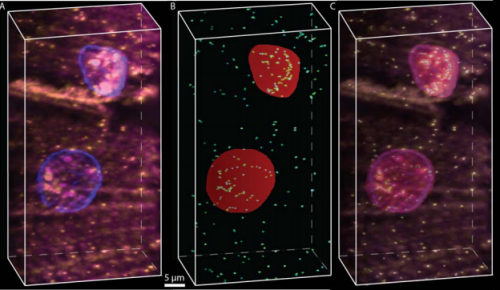
Single-molecule FISH in Drosophila muscle reveals location dependent mRNA composition of megaRNPs. Akiko Noma, Carlas S. Smith, Maximillian Huisman, Robert M. Martin, Melissa J. Moore, David Grunwald
Correlated Light-Serial Scanning Electron Microscopy (CoLSSEM) for ultrastructural visualization of single neurons in vivo. Yusuke Hirabayashi, Juan Carlos Tapia, Franck Polleux
3D clustering analysis of super-resolution microscopy data by 3D Voronoi tessellations. Leonid Andronov, Jonathan Michalon, Khalid Ouararhni, Igor Orlov, Ali Hamiche, Jean-Luc Vonesch, Bruno Klaholz
A simple open-source method for highly multiplexed imaging of single cells in tissues and tumours. Jia-Ren Lin, Benjamin Izar, Shaolin Mei, Shu Wang, Parin Shah, Peter Sorger
Morphometrics of complex cell shapes: Lobe Contribution Elliptic Fourier Analysis (LOCO-EFA). Yara E. Sanchez-Corrales, Matthew Hartley, Jop van Rooij, Stan F. M. Maree, Veronica Grieneisen
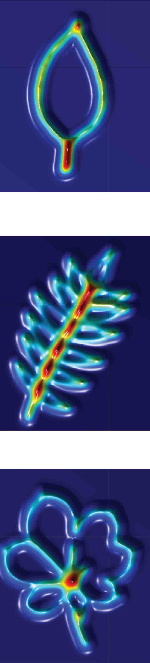
Persistent homology demarcates a leaf morphospace. Mao Li, Hong An, Ruthie Angelovici, Clement Bagaza, Albert Batushansky, Lynn Clark, Viktoriya Coneva, Michael Donoghue, Erika Edwards, Diego Fajardo, Hui Fang, Margaret Frank, Timothy Gallaher, Sarah Gebken, Theresa Hill, Shelley Jansky, Baljinder Kaur, Philip Klahs, Laura Klein, Vasu Kuraparthy, Jason Londo, Zoe Migicovsky, Allison Miller, Rebekah Mohn, Sean Myles, Wagner Otoni, J. Chris Pires, Edmond Riffer, Sam Schmerler, Elizabeth Spriggs, Christopher Topp, Allen Van Deynze, Kuang Zhang, Linglong Zhu, Braden M. Zink, Daniel H. Chitwood
Combining semi-automated image analysis techniques with machine learning algorithms to accelerate large scale genetic studies. Jonathan A. Atkinson, Guillaume Lobet, Manuel Noll, Patrick E. Meyer, Marcus Griffiths, Darren M. Wells
Near-Atomic Resolution Structure Determination in Over-Focus with Volta Phase Plate by Cs-corrected Cryo-EM. Xiao Fan, Lingyun Zhao, Chuan Liu, Jin-Can Zhang, Kelong Fan, Xiyun Yan, Hai-Lin Peng, Jianlin Lei, Hong-Wei Wang
A Guide To Analysis And Reconstruction Of Serial Block Face Scanning Electron Microscopy Data. Erin Cocks, Michael Taggart, Claire Rind, Kathryn White
Reflective imaging improves resolution, speed, and collection efficiency in light sheet microscopy. Yicong Wu, Abhishek Kumar, Corey Smith, Evan Ardiel, Panagiotis Chandris, Ryan Christensen, Ivan Rey-Suarez, Min Guo, Harshad Vishwasrao, Jiji Chen, Jianyong Tang, Arpita Upadhyaya, Patrick La Riviere, Hari Shroff
Molecular Recognition of Dopamine with Dual Near Infrared Excitation-Emission Two-Photon Microscopy. Jackson Travis Del Bonis-O’Donnell, Ralph H. Page, Abraham G. Beyene, Eric G. Tindall, Ian R. McFarlane, Markita P. Landry
Hemodynamic forces can be accurately measured in vivo with optical tweezers. Sebastien Harlepp, Fabrice Thalmann, Gautier Follain, Jacky G. Goetz
Polarization effects and the calibration of a donut beam axial optical tweezers for single-molecule force spectroscopy. Russell Pollari, Joshua N. Milstein
Measuring Protein Binding to Lipid Vesicles by Fluorescence Cross-Correlation Spectroscopy. Daniela Kruger, Jan Ebenhan, Kirsten Bacia
Controlling cell shape on hydrogels using lift-off patterning. Jens Moeller, Aleksandra K. Denisin, Joo Yong Sim, Robin E. Wilson, Alexandre J.S. Ribeiro, Beth L. Pruitt
A vacuum-actuated microtissue stretcher for long-term exposure to oscillatory strain within a 3D matrix. Matthew J. Walker, Michel Godin, Andrew Pelling
Universal Microfluidic System for Analysis and Control of Cell Dynamics. Ce Zhang, Hsiung-Lin Tu, Gengjie Jia, Tanzila Mukhtar, Verdon Taylor, Andrey Rzhetsky, Savas Tay
| Genome tools
The experimental design and data interpretation in ‘Unexpected mutations after CRISPR Cas9 editing in vivo’ by Schaefer et al. are insufficient to support the conclusions drawn by the authors. Christopher J. Wilson, Tim Fennell, Anne Bothmer, Morgan L. Maeder, Deepak Reyon, Cecilia Cotta-Ramusino, Cecilia A. Fernandez, Eugenio Marco, Luis A. Barrera, Hariharan Jayaram, Charles F. Albright, Gerald F. Cox, George M. Church, Vic E. Myer
Questioning unexpected CRISPR off-target mutations in vivo. Sang-Tae Kim, Jeongbin Park, Daesik Kim, Kyoungmi Kim, Sangsu Bae, Matthias Schlesner, Jin-Soo Kim
Response to Editas: Unexpected mutations after CRISPR-Cas9 editing in vivo. Kellie A. Schaefer, Wen-Hsuan Wu, Diana F. Colgan, Stephen H. Tsang, Alexander G. Bassuk, Vinit B. Mahajan
mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces potentially functional transcripts. Jennifer L. Anderson, Timothy S. Mulligan, Meng-Chieh Shen, Hui Wang, Catherine M. Scahill, Shao J. Du, Elisabeth M. Busch-Nentwich, Steven A. Farber
Correction of copy number induced false positives in CRISPR screens. Antoine de Weck, Javad Golji, Michael D. Jones, Joshua M. Korn, Eric Billy, E. Robert McDonald III, Tobias Schmelzle, Hans Bitter, Audrey Kauffmann
CRISPRi is not strand-specific and redefines the transcriptional landscape. Françoise S. Howe, Andrew Russell, Afaf El-Sagheer, Anitha Nair, Tom Brown, Jane Mellor
Inducible, tunable and multiplex human gene regulation using CRISPR-Cpf1-based transcription factors. Yu Gyoung Tak, Benjamin P. Kleinstiver, James K. Nuñez, Jonathan Y. Hsu, Jingyi Gong, Jonathan S. Weissman, J. Keith Joung
CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Miguel A. Moreno-Mateos, Juan P. Fernandez, Romain Rouet, Maura A. Lane, Charles E. Vejnar, Emily Mis, Mustafa K. Khokha, Jennifer A. Doudna, Antonio J. Giraldez
Highly parallel genome variant engineering with CRISPR/Cas9 in eukaryotic cells. Meru J. Sadhu, Joshua S. Bloom, Laura Day, Jake J. Siegel, Sriram Kosuri, Leonid Kruglyak
Germline Cas9 Expression Yields Highly Efficient Genome Engineering in a Major Worldwide Disease Vector, Aedes aegypti. Ming Li, Michelle Bui, Ting Yang, Bradley White, Omar Akbari
Highly efficient site-specific mutagenesis in Malaria mosquitoes using CRISPR. Ming Li, Omar Akbari, Bradley White
Reducing resistance allele formation in CRISPR gene drives. Jackson Champer, Jingxian Liu, Suh Yeon Oh, Riona Reeves, Anisha Luthra, Nathan Oakes, Andrew G. Clark,Philipp W. Messer
Engineering Cell Sensing and Responses Using a GPCR-Coupled CRISPR-Cas System. P. C. Dave P. Dingal, Nathan H. Kipniss, Louai Labanieh, Yuchen Gao, Lei S. Qi
TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Lexiang Ji, William T. Jordan, Xiuling Shi, Lulu Hu, Chuan He, Robert J. Schmitz
Addressing the looming identity crisis in single cell RNA-seq. Megan Crow, Anirban Paul, Sara Ballouz, Z. Josh Huang, Jesse Gillis
Ultrafast comparison of personal genomes. Gustavo Glusman, Denise E. Mauldin, Leroy E. Hood, Max Robinson
Critical Assessment of Metagenome Interpretation − a benchmark of computational metagenomics software. Alexander Sczyrba, Peter Hofmann, Peter Belmann, David Koslicki, Stefan Janssen, Johannes Droege, Ivan Gregor, Stephan Majda, Jessika Fiedler, Eik Dahms, Andreas Bremges, Adrian Fritz, Ruben Garrido-Oter, Tue Sparholt Jorgensen, Nicole Shapiro, Philip D Blood, Alexey Gurevich, Yang Bai, Dmitrij Turaev, Matthew Z DeMaere, Rayan Chikhi, Niranjan Nagarajan, Christopher Quince, Fernando Meyer, Monika Balvociute, Lars Hestbjerg Hansen, Soren J Sorensen, Burton K H Chia, Bertrand Denis, Jeff L Froula, Zhong Wang, Robert Egan, Dongwan Don Kang, Jeffrey J Cook, Charles Deltel, Michael Beckstette, Claire Lemaitre, Pierre Peterlongo, Guillaume Rizk, Dominique Lavenier, Yu-Wei Wu, Steven W Singer, Chirag Jain, Marc Strous, Heiner Klingenberg, Peter Meinicke, Michael Barton, Thomas Lingner, Hsin-Hung Lin, Yu-Chieh Liao, Genivaldo Gueiros Z Silva, Daniel A Cuevas, Robert A Edwards, Surya Saha, Vitor C Piro, Bernhard Y Renard, Mihai Pop, Hans-Peter Klenk, Markus Goeker, Nikos C Kyrpides, Tanja Woyke, Julia A Vorholt, Paul Schulze-Lefert, Edward M Rubin, Aaron E Darling, Thomas Rattei, Alice C McHardy
Second-generation method for analysis of chromatin binding using formaldehyde crosslinking kinetics. Hussain Zaidi, Elizabeth A. Hoffman, Savera J. Shetty, Stefan Bekiranov, David T. Auble
Identification and visualization of differential isoform expression in RNA-seq time series. Maria Jose Nueda, Jordi Martorell-Marugan, Cristina Marti, Sonia Tarazona, Ana Conesa
High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Todd P. Michael, Florian Jupe, Felix Bemm, Stanley T. Motley, Justin P. Sandoval, Olivier Loudet, Detlef Weigel, Joseph R. Ecker
Parallel Computing to Speed up Whole-Genome Bayesian Regression Analyses Using Orthogonal Data Augmentation. Hao Cheng, Rohan Fernando, Dorian Garrick
| Resources
scmap – A tool for unsupervised projection of single cell RNA-seq data. Vladimir Yu. Kiselev, Martin Hemberg
xCell: Digitally portraying the tissue cellular heterogeneity landscape. Dvir Aran, Zicheng Hu, Atul J Butte
DeepSF: deep convolutional neural network for mapping protein sequences to folds. Jie Hou, Badri Adhikari, Jianlin Cheng
A robust statistical framework to detect multiple sources of hidden variation in single-cell transcriptomes. Donghyung Lee, Anthony Cheng, Duygu Ucar
MIMoSA: A Method for Inter-Modal Segmentation Analysis. Alessandra Valcarcel, Kristin Linn, Simon Vandekar, Theodore Satterthwaite, Peter Calabresi, Dzung Pham, Russell Shinohara
scImpute: Accurate And Robust Imputation For Single Cell RNA-Seq Data. Wei Vivian Li, Jingyi Jessica Li
Inter-Tools: a toolkit for interactome analysis. Hannah Beth Catabia, Caren Smith, Jose Ordovas
GARFIELD-NGS: Genomic vARiants FIltering by dEep Learning moDels in NGS. Viola Ravasio, Edoardo Giacopuzzi
PinAPL-Py: a web-service for the analysis of CRISPR-Cas9 Screens. Philipp N Spahn, Tyler Bath, Ryan J. Weiss, Jihoon Kim, Jeffrey D. Esko, Nathan E Lewis, Olivier Harismendy
GrapHi-C: Graph-based visualization of Hi-C Datasets. Kimberly MacKay, Anthony Kusalik, Christopher H. Eskiw
IndeCut evaluates performance of network motif discovery algorithms. Mitra Ansariola, Molly Megraw, David Koslicki
ADAGE signature analysis: differential expression analysis with data-defined gene sets. Jie Tan, Matthew Huyck, Dongbo Hu,Rene A. Zelaya,Deborah A. Hogan, Casey S. Greene
MoMo: Discovery of post-translational modification motifs. Alice Cheng, Charles Grant, Timothy L. Bailey, William Noble
TiSAn: Tissue Specific Variant Annotation. Kevin Vervier, Jacob J. Michaelson
Patternize: An R Package For Quantifying Color Pattern Variation. Steven M. Van Belleghem, Riccardo Papa, Humberto Ortiz-Zuazaga, Frederik Hendrickx, Chris Jiggins, W. Owen McMillan, Brian Counterman
powsimR: Power analysis for bulk and single cell RNA-seq experiments. Beate Vieth, Christoph Ziegenhain, Swati Parekh, Wolfgang Enard, Ines Hellmann
GIGGLE: a search engine for large-scale integrated genome analysis. Ryan M. Layer, Brent S. Pedersen, Tonya DiSera, Gabor T. Marth, Jason Gertz, Aaron R. Quinlan
Accessible, curated metagenomic data through ExperimentHub. Edoardo Pasolli, Lucas Schiffer, Paolo Manghi, Audrey Renson, Valerie Obenchain, Duy Tin Truong, Francesco Beghini, Faizan Malik, Marcel Ramos, Jennifer B Dowd, Curtis Huttenhower, Martin Morgan, Nicola Segata, Levi Waldron
EMHP: An accurate automated hole masking algorithm for single-particle cryo-EM image processing. Zachary Berndsen, Charles Bowman Jr., Haerin Jang, Andrew B. Ward
Ludicrous Speed Linear Mixed Models for Genome-Wide Association Studies. Carl M. Kadie, David Heckerman
Research practice
A mathematical theory of knowledge, science, bias and pseudoscience. Daniele Fanelli .
The earth is flat (p>0.05): Significance thresholds and the crisis of unreplicable research. Valentin Amrhein, Fränzi Korner-Nievergelt, Tobias Roth
Can editors protect peer review from bad reviewers? Rafael D’Andrea, James P O’Dwyer
Transitioning “Open Data” from a NOUN to a VERB. Aadinarayana varma D, Sheevendra Sharma
Examining publication bias – A simulation-based evaluation of statistical tests on publication bias. Andreas Schneck
Not All Experimental Questions Are Created Equal: Accelerating Biological Data to Knowledge Transformation (BD2K) via Science Informatics, Active Learning and Artificial Intelligence. Simon Kasif, Stanley Letovsky, Richard J. Roberts, Martin Steffen
A guide to robust statistical methods in neuroscience. Rand R. Wilcox, Guillaume A. Rousselet
Experimenting with reproducibility in bioinformatics.Yang-Min Kim, Jean-Baptiste Poline, Guillaume Dumas
Online, Interactive Tutorials Provide Effective Instruction in Science Process Skills. Maxwell Kramer, Dalay Olson, J.D. Walker
Why not…
Modelling personality, plasticity and predictability in shelter dogs. Conor Goold, Ruth C. Newberry
An outbreak of Pseudomonas aeruginosa infection linked to a Black Friday piercing event. Peter MacPherson, Katherine Valentine, Victoria Chadderton, Evdokia Dardamissis, Ian Doige, Andrew Fox, Sam Ghebrehewet, Thomas Hampton, Ken Mutton, Claire Sherratt, Catherine M. McCann


 (No Ratings Yet)
(No Ratings Yet)